3GXB
 
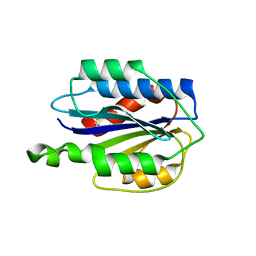 | | Crystal structure of VWF A2 domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | 著者 | Zhou, Y.F, Springer, T.A. | | 登録日 | 2009-04-02 | | 公開日 | 2009-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von Willebrand factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4M5E
 
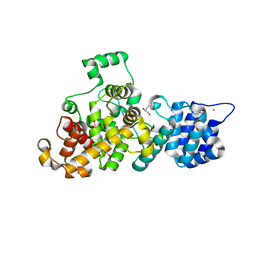 | | Tse3 structure | | 分子名称: | CADMIUM ION, CALCIUM ION, GLYCEROL, ... | | 著者 | Qian, Y. | | 登録日 | 2013-08-08 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
5G4C
 
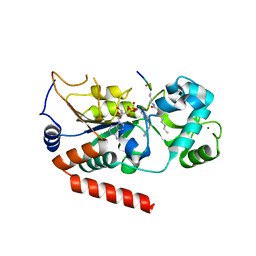 | | Human SIRT2 catalyse short chain fatty acyl lysine | | 分子名称: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2, SIRT2, ... | | 著者 | Wang, Y. | | 登録日 | 2016-05-09 | | 公開日 | 2017-05-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | SIRT2 Reverses 4-Oxononanoyl Lysine Modification on Histones.
J. Am. Chem. Soc., 138, 2016
|
|
226L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
223L
 
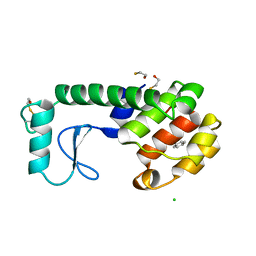 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
222L
 
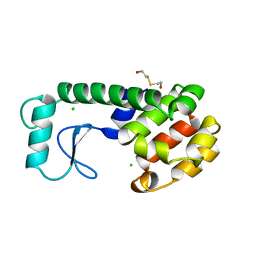 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
1LBA
 
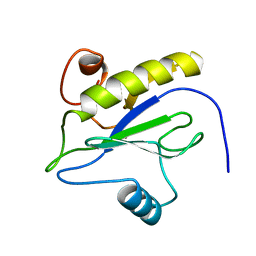 | |
5HZ5
 
 | |
252L
 
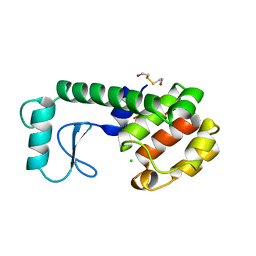 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-10-28 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
2JUS
 
 | |
229L
 
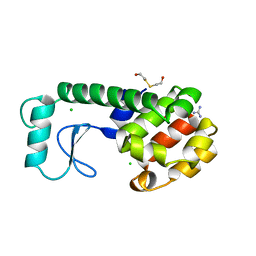 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GUANIDINE, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-26 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
227L
 
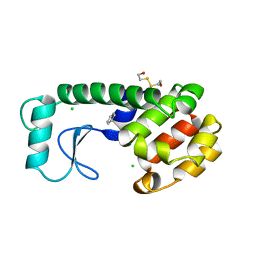 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
2H2H
 
 | |
4L23
 
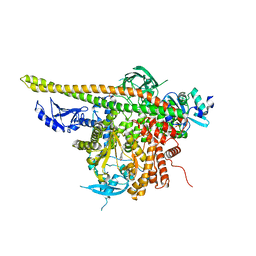 | | Crystal Structure of p110alpha complexed with niSH2 of p85alpha and PI-103 | | 分子名称: | 3-(4-MORPHOLIN-4-YLPYRIDO[3',2':4,5]FURO[3,2-D]PYRIMIDIN-2-YL)PHENOL, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | 著者 | Zhang, J, Zhao, Y.L, Chen, Y.Y, Huang, M, Jiang, F. | | 登録日 | 2013-06-04 | | 公開日 | 2014-01-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
ACS Med Chem Lett, 5, 2014
|
|
2JSZ
 
 | |
2NB4
 
 | | Solution structure of Q388A3 PDZ domain | | 分子名称: | Putative uncharacterized protein | | 著者 | Mei, S. | | 登録日 | 2016-01-24 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Q388A3 PDZ domain from Trypanosoma brucei
J.Struct.Biol., 194, 2016
|
|
2N7S
 
 | |
8J6S
 
 | | Cryo-EM structure of the single CAF-1 bound right-handed Di-tetrasome | | 分子名称: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | 著者 | Liu, C.P, Yu, Z.Y, Xu, R.M. | | 登録日 | 2023-04-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8J6T
 
 | | Cryo-EM structure of the double CAF-1 bound right-handed Di-tetrasome | | 分子名称: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | 著者 | Liu, C.P, Yu, Z.Y, Xu, R.M. | | 登録日 | 2023-04-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8A2G
 
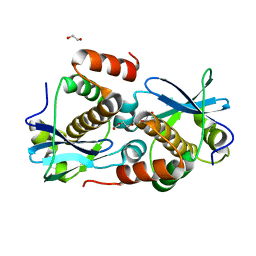 | | Crystal structure of Sebokelevirus 2A2 protein | | 分子名称: | 1,2-ETHANEDIOL, 2A2 protein, TETRAETHYLENE GLYCOL | | 著者 | Zhu, L, Von Castelmur, E, Whang, X, Ren, J, Fry, E, Perrakis, A, Stuart, D.I. | | 登録日 | 2022-06-03 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Structural plasticity of 2A proteins in the Parechovirus family
to be published
|
|
7BH2
 
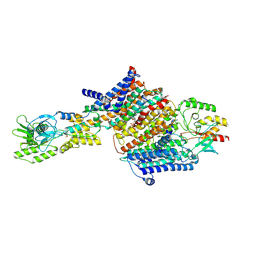 | | Cryo-EM Structure of KdpFABC in E2Pi state with BeF3 and K+ | | 分子名称: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | 著者 | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | 登録日 | 2021-01-09 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BPC
 
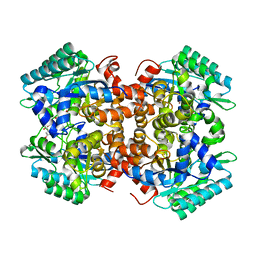 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum in complex with 2,5-DHBA | | 分子名称: | 2,3-dihydroxybenzoate decarboxylase, 2,5-dihydroxybenzoic acid, ZINC ION | | 著者 | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | 登録日 | 2020-03-22 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
7BH1
 
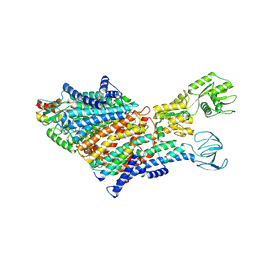 | | Cryo-EM Structure of KdpFABC in E1 state with K | | 分子名称: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | 著者 | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | 登録日 | 2021-01-09 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BGY
 
 | | Cryo-EM Structure of KdpFABC in E2Pi state with MgF4 | | 分子名称: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, MAGNESIUM ION, ... | | 著者 | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | 登録日 | 2021-01-09 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BP1
 
 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum in complex with Catechol | | 分子名称: | 2,3-dihydroxybenzoate decarboxylase, CATECHOL, ZINC ION | | 著者 | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | 登録日 | 2020-03-21 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
