7XLB
 
 | |
6KIG
 
 | | Structure of cyanobacterial photosystem I-IsiA supercomplex | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | 登録日 | 2019-07-18 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-03-04 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
4EIW
 
 | | Whole cytosolic region of atp-dependent metalloprotease FtsH (G399L) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH | | 著者 | Suno, R, Niwa, H, Tsuchiya, D, Yoshida, M, Morikawa, K. | | 登録日 | 2012-04-06 | | 公開日 | 2012-06-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Structure of the whole cytosolic region of ATP-dependent protease FtsH
Mol.Cell, 22, 2006
|
|
5HAV
 
 | |
5WY2
 
 | |
5ZND
 
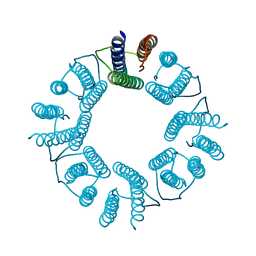 | | 8-mer nanotube derived from 24-mer rHuHF nanocage | | 分子名称: | Ferritin heavy chain | | 著者 | Wang, W.M, Wang, L.L, Zang, J.C, Chen, H, Zhao, G.H, Wang, H.F. | | 登録日 | 2018-04-09 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Selective Elimination of the Key Subunit Interfaces Facilitates Conversion of Native 24-mer Protein Nanocage into 8-mer Nanorings.
J. Am. Chem. Soc., 140, 2018
|
|
7YA1
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9S
 
 | | Cryo-EM structure of apo SARS-CoV-2 Omicron spike protein (S-2P-GSAS) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-26 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
2I89
 
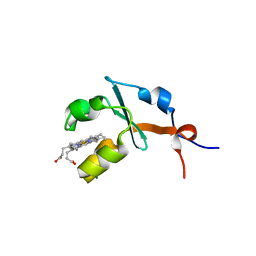 | | Structure of septuple mutant of Rat Outer Mitochondrial Membrane Cytochrome B5 | | 分子名称: | Cytochrome b5 type B, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Terzyan, S, Zhang, X.C, Benson, D.R, Wang, L, Sun, N. | | 登録日 | 2006-09-01 | | 公開日 | 2006-10-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A histidine/tryptophan pi-stacking interaction stabilizes the heme-independent folding core of microsomal apocytochrome b5 relative to that of mitochondrial apocytochrome b5.
Biochemistry, 45, 2006
|
|
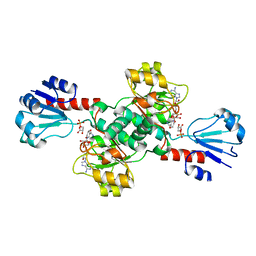
6CWA
 
 | |
4JYU
 
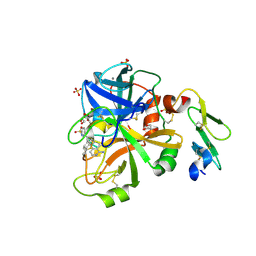 | |
4JYV
 
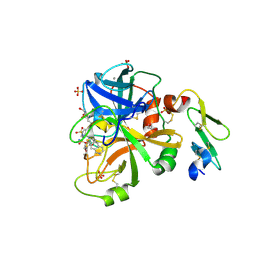 | |
4KBS
 
 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 12:0 phosphatidic acid (12:0 PA) | | 分子名称: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-ETHANEDIOL, Glycolipid transfer protein domain-containing protein 1 | | 著者 | Simanshu, D.K, Brown, R.E, Patel, D.J. | | 登録日 | 2013-04-23 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.898 Å) | | 主引用文献 | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
2N2N
 
 | |
8HPU
 
 | |
8HQ7
 
 | |
8HPF
 
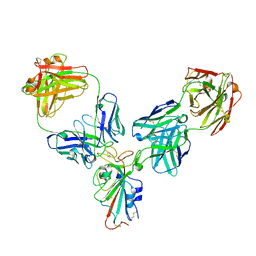 | |
8HPV
 
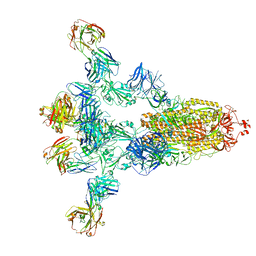 | |
7CGP
 
 | | Cryo-EM structure of the human mitochondrial translocase TIM22 complex at 3.7 angstrom. | | 分子名称: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, Acylglycerol kinase, mitochondrial, ... | | 著者 | Qi, L, Wang, Q, Guan, Z, Yan, C, Yin, P. | | 登録日 | 2020-07-01 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-03-17 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of the human mitochondrial translocase TIM22 complex.
Cell Res., 31, 2021
|
|
7CKA
 
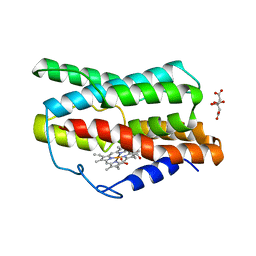 | |
6IF4
 
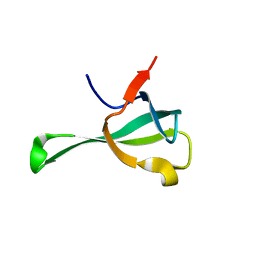 | | Crystal structure of Tbtudor | | 分子名称: | Histone acetyltransferase | | 著者 | Gao, J, Ye, K, Diwu, Y, Liao, S, Tu, X. | | 登録日 | 2018-09-18 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.934 Å) | | 主引用文献 | Crystal structure of TbEsa1 presumed Tudor domain from Trypanosoma brucei.
J.Struct.Biol., 209, 2020
|
|
2N31
 
 | |
3RKD
 
 | |
3K1Q
 
 | | Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics | | 分子名称: | Core protein VP6, Outer capsid VP5, Outer capsid VP7, ... | | 著者 | Cheng, L.P, Zhu, J, Hiu, W.H, Zhang, X.K, Honig, B, Fang, Q, Zhou, Z.H. | | 登録日 | 2009-09-28 | | 公開日 | 2010-03-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Backbone Model of an Aquareovirus Virion by Cryo-Electron Microscopy and Bioinformatics
J.Mol.Biol., 397, 2010
|
|
7XCK
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 fab (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, S309 light chain, ... | | 著者 | Gao, G.F, Qi, J.X, Zhao, Z.N, Xie, Y.F, Liu, S. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
