7W6I
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me1 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W67
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me0 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.194 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6J
 
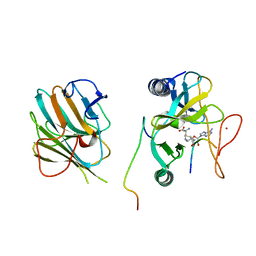 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me2 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7E2U
 
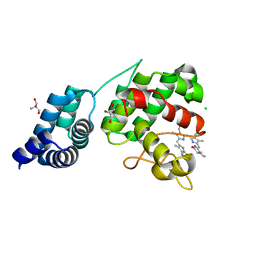 | | Synechocystis GUN4 in complex with phytochrome | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[5-[(~{Z})-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[(~{Z})-[5-[(~{Z})-[(4~{R})-3-ethyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, CHLORIDE ION, ... | | 著者 | Liu, L, Hu, J. | | 登録日 | 2021-02-07 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
7E2T
 
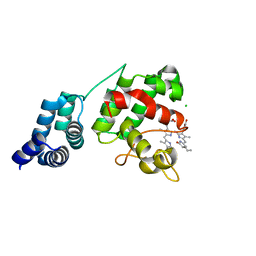 | | Synechocystis GUN4 in complex with phycocyanobilin | | 分子名称: | 3-[2-[(~{Z})-[5-[(~{Z})-[(4~{R})-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, CHLORIDE ION, Ycf53-like protein | | 著者 | Liu, L, Hu, J. | | 登録日 | 2021-02-07 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
7E2R
 
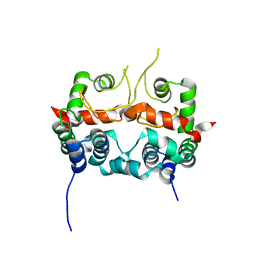 | |
7E2S
 
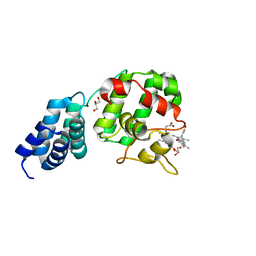 | |
8A2F
 
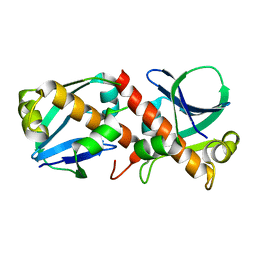 | | Crystal Structure of Ljunganvirus 1 2A protein | | 分子名称: | 2A protein | | 著者 | von Castelmur, E, Zhu, L, Wang, X, Fry, E, Ren, J, Perrakis, A, Stuart, D.I. | | 登録日 | 2022-06-03 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Structural plasticity of 2A proteins in the Parechovirus family
To Be Published
|
|
8A2E
 
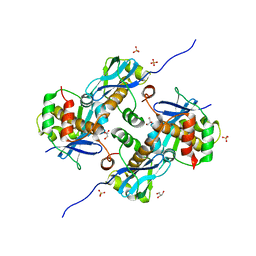 | | Crystal Structure of Human Parechovirus 3 2A protein | | 分子名称: | 2A protein, GLYCEROL, SULFATE ION | | 著者 | von Castelmur, E, Zhu, L, wang, X, Fry, E, Ren, J, Perrakis, A, Stuart, D.I. | | 登録日 | 2022-06-03 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural plasticity of 2A proteins in the Parechovirus family.
To Be Published
|
|
7FC3
 
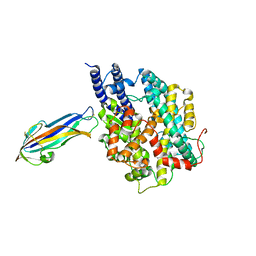 | | structure of NL63 receptor-binding domain complexed with horse ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | 著者 | Wang, X.Q, Ge, J.W, Lan, J. | | 登録日 | 2021-07-13 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
7EUO
 
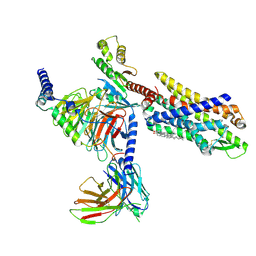 | | The structure of formyl peptide receptor 1 in complex with Gi and peptide agonist fMLF | | 分子名称: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, X.K, Chen, G, Liao, Q.W, Du, Y, Hu, H.L, Ye, D.Q. | | 登録日 | 2021-05-18 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for recognition of N-formyl peptides as pathogen-associated molecular patterns.
Nat Commun, 13, 2022
|
|
7FC5
 
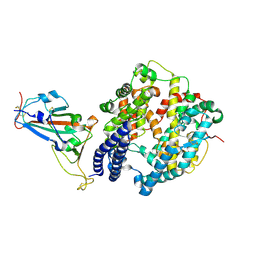 | | Crystal structure of SARS-CoV-2 RBD and horse ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1 | | 著者 | Wang, X.Q, Lan, J, Ge, J.W. | | 登録日 | 2021-07-13 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.894 Å) | | 主引用文献 | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
7FC6
 
 | | Crystal structure of SARS-CoV RBD and horse ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | 著者 | Wang, X.Q, Lan, J, Ge, J.W. | | 登録日 | 2021-07-13 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.655 Å) | | 主引用文献 | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
7E9T
 
 | | Nanometer resolution in situ structure of SARS-CoV-2 post-fusion spike | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2, ... | | 著者 | Zhu, Y, Tai, L, Zhu, G, Yin, G, Sun, F. | | 登録日 | 2021-03-05 | | 公開日 | 2021-11-17 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (10.9 Å) | | 主引用文献 | Nanometer-resolution in situ structure of the SARS-CoV-2 postfusion spike protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3RDN
 
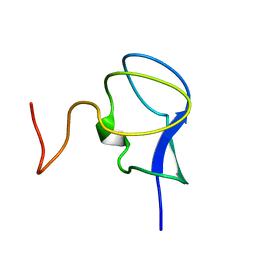 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | ANTIFREEZE PROTEIN RD3 TYPE III | | 著者 | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | 登録日 | 1998-02-24 | | 公開日 | 1999-02-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
7EV8
 
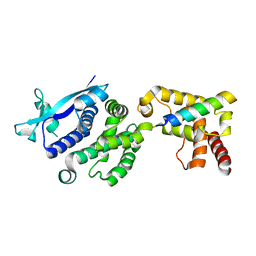 | |
7CT5
 
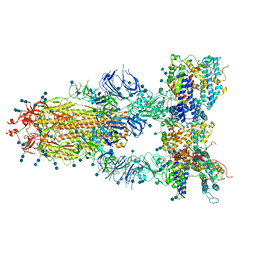 | | S protein of SARS-CoV-2 in complex bound with T-ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Guo, L, Bi, W.W, Zhang, Y.Y, Yan, R.H, Li, Y.N, Zhou, Q, Dang, B.B. | | 登録日 | 2020-08-18 | | 公開日 | 2020-11-18 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Engineered trimeric ACE2 binds viral spike protein and locks it in "Three-up" conformation to potently inhibit SARS-CoV-2 infection.
Cell Res., 31, 2021
|
|
7C2E
 
 | | GLP-1R-Gs complex structure with a small molecule full agonist | | 分子名称: | 2-[[4-[6-[(4-cyano-2-fluoranyl-phenyl)methoxy]pyridin-2-yl]-3,6-dihydro-2~{H}-pyridin-1-yl]methyl]-3-[[(2~{S})-oxetan-2-yl]methyl]imidazo[4,5-b]pyridine-5-carboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Ma, H, Yuan, D.P, Huang, W, Wenge, Z, Xu, E. | | 登録日 | 2020-05-07 | | 公開日 | 2020-08-26 | | 最終更新日 | 2020-12-16 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural insights into the activation of GLP-1R by a small molecule agonist.
Cell Res., 30, 2020
|
|
3NLA
 
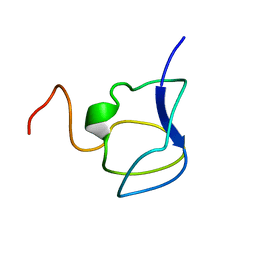 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, 40 STRUCTURES | | 分子名称: | ANTIFREEZE PROTEIN RD3 TYPE III | | 著者 | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | 登録日 | 1998-02-24 | | 公開日 | 1999-02-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
4K0U
 
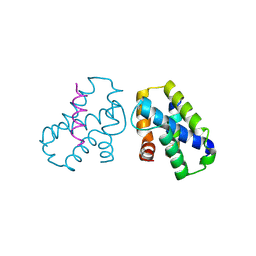 | | Pilotin/secretin peptide Complex | | 分子名称: | Lipoprotein OutS, Type II secretion system protein D | | 著者 | Rehman, S, Pickersgill, R.W. | | 登録日 | 2013-04-04 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Anatomy of secretin binding to the Dickeya dadantii type II secretion system pilotin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8HAW
 
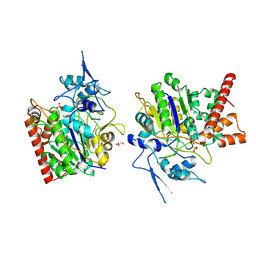 | | An auto-activation mechanism of plant non-specific phospholipase C | | 分子名称: | CALCIUM ION, GLYCEROL, Non-specific phospholipase C4, ... | | 著者 | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | 登録日 | 2022-10-26 | | 公開日 | 2023-01-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
8HAV
 
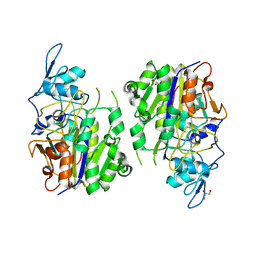 | | An auto-activation mechanism of plant non-specific phospholipase C | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Non-specific phospholipase C4 | | 著者 | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | 登録日 | 2022-10-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
7WPP
 
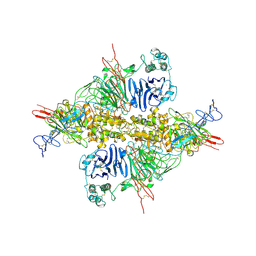 | | Cryo-EM structure of VWF D'D3 dimer complexed with D1D2 at 2.85 angstron resolution (1 unit) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | 著者 | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | 登録日 | 2022-01-24 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-06-15 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
7WPQ
 
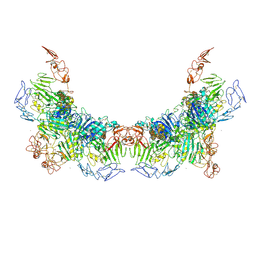 | | Cryo-EM structure of VWF D'D3 dimer complexed with D1D2 at 3.27 angstron resolution (2 units) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | 著者 | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | 登録日 | 2022-01-24 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-06-15 | | 実験手法 | ELECTRON MICROSCOPY (3.267 Å) | | 主引用文献 | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
7WQT
 
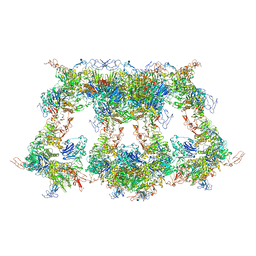 | | Cryo-EM structure of VWF D'D3 dimer complexed with D1D2 at 4.3 angstron resolution (VWF tube) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | 著者 | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | 登録日 | 2022-01-26 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-06-15 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
