4QL5
 
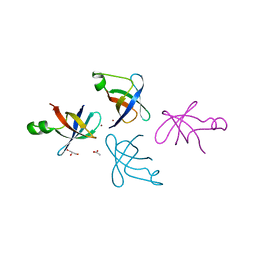 | | Crystal structure of translation initiation factor IF-1 from Streptococcus pneumoniae TIGR4 | | 分子名称: | ACETATE ION, GLYCEROL, Translation initiation factor IF-1, ... | | 著者 | Stogios, P.J, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-06-10 | | 公開日 | 2014-07-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.025 Å) | | 主引用文献 | Crystal structure of translation initiation factor IF-1 from Streptococcus pneumoniae TIGR4
TO BE PUBLISHED
|
|
4IC1
 
 | | Crystal structure of SSO0001 | | 分子名称: | IRON/SULFUR CLUSTER, MANGANESE (II) ION, Uncharacterized protein | | 著者 | Nocek, B, Skarina, T, Lemak, S, Beloglazova, N, Flick, R, Brown, G, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-12-09 | | 公開日 | 2013-01-16 | | 最終更新日 | 2014-07-02 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus.
J.Am.Chem.Soc., 135, 2013
|
|
4WZ3
 
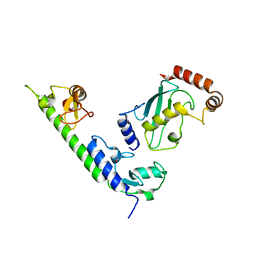 | | Crystal structure of the complex between LubX/LegU2/Lpp2887 U-box 1 and Homo sapiens UBE2D2 | | 分子名称: | E3 ubiquitin-protein ligase LubX, Ubiquitin-conjugating enzyme E2 D2 | | 著者 | Stogios, P.J, Quaile, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-18 | | 公開日 | 2015-01-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
4R01
 
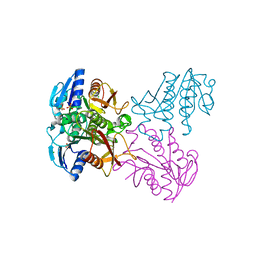 | | Crystal structure of SP1627, a putative NADH-flavin reductase, from Streptococcus pneumoniae TIGR4 | | 分子名称: | CHLORIDE ION, SULFATE ION, putative NADH-flavin reductase | | 著者 | Stogios, P.J, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-07-29 | | 公開日 | 2014-08-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of SP1627, a putative NADH-flavin reductase, from Streptococcus pneumoniae TIGR4
TO BE PUBLISHED
|
|
4RO3
 
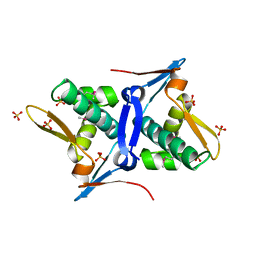 | | 1.8 Angstrom Crystal Structure of the N-terminal Domain of Protein with Unknown Function from Vibrio cholerae. | | 分子名称: | Hypothetical Protein, SULFATE ION | | 著者 | Minasov, G, Wawrzak, Z, Stogios, P.J, Skarina, T, Seed, K.D, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-10-27 | | 公開日 | 2014-12-03 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 1.8 Angstrom Crystal Structure of the N-terminal Domain of Protein with Unknown Function from Vibrio cholerae.
To be Published
|
|
4WZ2
 
 | | Crystal structure of U-box 2 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris, Ile175Met mutant | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase LubX, HEXANE-1,6-DIOL | | 著者 | Stogios, P.J, Qualie, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-11-18 | | 公開日 | 2015-01-28 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (3.408 Å) | | 主引用文献 | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
7S2I
 
 | | Crystal structure of sulfonamide resistance enzyme Sul1 in complex with 6-hydroxymethylpterin | | 分子名称: | 6-HYDROXYMETHYLPTERIN, CHLORIDE ION, GLYCEROL, ... | | 著者 | Stogios, P.J, Skarina, T, Kim, Y, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-09-03 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2J
 
 | | Crystal structure of sulfonamide resistance enzyme Sul2 apoenzyme | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Stogios, P.J, Skarina, T, Michalska, K, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-09-03 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2M
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 in complex with 6-hydroxymethylpterin | | 分子名称: | 6-HYDROXYMETHYLPTERIN, Sul3 | | 著者 | Stogios, P.J, Skarina, T, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-09-03 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2K
 
 | | Crystal structure of sulfonamide resistance enzyme Sul2 in complex with 7,8-dihydropteroate, magnesium, and pyrophosphate | | 分子名称: | 4-AMINOBENZOIC ACID, 7,8-DIHYDROPTEROATE, CHLORIDE ION, ... | | 著者 | Stogios, P.J, Skarina, T, Michalska, K, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-09-03 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2L
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 apoenzyme | | 分子名称: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | 著者 | Stogios, P.J, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-09-03 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
6BAL
 
 | | 2.1 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae in Complex with L-Malate | | 分子名称: | (2S)-2-hydroxybutanedioic acid, CHLORIDE ION, Malate dehydrogenase | | 著者 | Minasov, G, Wawrzak, Z, Skarina, T, Grimshaw, S, Satchell, K.J.F, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-10-13 | | 公開日 | 2017-10-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | 2.1 Angstrom Resolution Crystal Structure of Malate Dehydrogenase from Haemophilus influenzae in Complex with L-Malate
To Be Published
|
|
6B8W
 
 | | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae. | | 分子名称: | MANGANESE (II) ION, THIOCYANATE ION, XRE family transcriptional regulator | | 著者 | Minasov, G, Wawrzak, Z, Skarina, T, McChesney, C, Grimshaw, S, Sandoval, J, Satchell, K.J.F, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-10-09 | | 公開日 | 2017-10-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae.
To Be Published
|
|
6CZP
 
 | | 2.2 Angstrom Resolution Crystal Structure Oxygen-Insensitive NAD(P)H-dependent Nitroreductase NfsB from Vibrio vulnificus in Complex with FMN | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Minasov, G, Wawrzak, Z, Skarina, T, Grimshaw, S, Kwon, K, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-04-09 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | 2.2 Angstrom Resolution Crystal Structure Oxygen-Insensitive NAD(P)H-dependent Nitroreductase NfsB from Vibrio vulnificus in Complex with FMN.
To Be Published
|
|
7K3N
 
 | | Crystal Structure of NSP1 from SARS-CoV-2 | | 分子名称: | Host translation inhibitor nsp1 | | 著者 | Semper, C, Watanabe, N, Chang, C, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-09-11 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural characterization of nonstructural protein 1 from SARS-CoV-2.
Iscience, 24, 2021
|
|
6CN0
 
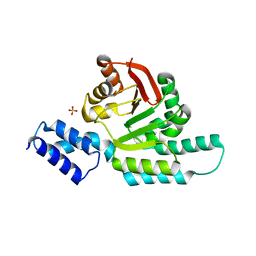 | | 2.95 Angstrom Crystal Structure of 16S rRNA Methylase from Proteus mirabilis | | 分子名称: | 16S rRNA (guanine(1405)-N(7))-methyltransferase, CHLORIDE ION, CITRIC ACID, ... | | 著者 | Minasov, G, Wawrzak, Z, Di Leo, R, Evdokimova, E, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-03-06 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | 2.95 Angstrom Crystal Structure of 16S rRNA Methylase from Proteus mirabilis.
To Be Published
|
|
6DKH
 
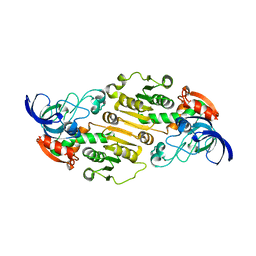 | | The crystal structure of L-idonate 5-dehydrogenase from Escherichia coli str. K-12 substr. MG1655 | | 分子名称: | L-idonate 5-dehydrogenase (NAD(P)(+)), ZINC ION | | 著者 | Tan, K, Evdokimova, E, McChesney, C, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-05-29 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.608 Å) | | 主引用文献 | The crystal structure of L-idonate 5-dehydrogenase from Escherichia coli str. K-12 substr. MG1655
To Be Published
|
|
1K77
 
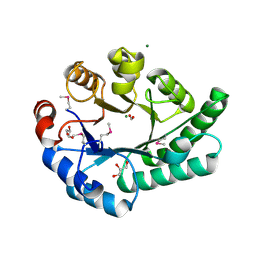 | | Crystal Structure of EC1530, a Putative Oxygenase from Escherichia coli | | 分子名称: | FORMIC ACID, GLYCEROL, Hypothetical protein ygbM, ... | | 著者 | Kim, Y, Skarina, T, Beasley, S, Laskowski, R, Arrowsmith, C.H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2001-10-18 | | 公開日 | 2002-03-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Crystal structure of Escherichia coli EC1530, a glyoxylate induced protein YgbM.
Proteins, 48, 2002
|
|
1KS2
 
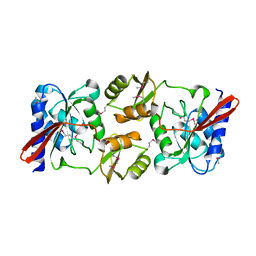 | | Crystal Structure Analysis of the rpiA, Structural Genomics, protein EC1268. | | 分子名称: | protein EC1268, RPIA | | 著者 | Zhang, R, Joachimiak, A, Edwards, A.M, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-01-10 | | 公開日 | 2002-08-14 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of Escherichia coli ribose-5-phosphate isomerase: a ubiquitous enzyme of the pentose phosphate pathway and the Calvin cycle.
STRUCTURE, 11, 2003
|
|
4KSN
 
 | |
4GYT
 
 | | Crystal structure of lpg0076 protein from Legionella pneumophila | | 分子名称: | SODIUM ION, uncharacterized protein | | 著者 | Michalska, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-09-05 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.047 Å) | | 主引用文献 | Crystal structure of lpg0076 protein from Legionella pneumophila
To be Published
|
|
1LXJ
 
 | | X-RAY STRUCTURE OF YBL001c NORTHEAST STRUCTURAL GENOMICS (NESG) CONSORTIUM TARGET YTYst72 | | 分子名称: | HYPOTHETICAL 11.5KDA PROTEIN IN HTB2-NTH2 INTERGENIC REGION, SULFATE ION | | 著者 | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2002-06-05 | | 公開日 | 2003-07-29 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | CRYSTAL STRUCTURES OF MTH1187 AND ITS YEAST ORTHOLOG YBL001C
Proteins, 52, 2003
|
|
3LQY
 
 | | Crystal structure of putative isochorismatase hydrolase from Oleispira antarctica | | 分子名称: | GLYCEROL, putative isochorismatase hydrolase | | 著者 | Goral, A, Chruszcz, M, Kagan, O, Cymborowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-02-10 | | 公開日 | 2010-03-16 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of a putative isochorismatase hydrolase from Oleispira antarctica.
J.Struct.Funct.Genom., 13, 2012
|
|
1LXN
 
 | | X-RAY STRUCTURE OF MTH1187 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT272 | | 分子名称: | HYPOTHETICAL PROTEIN MTH1187, SULFATE ION | | 著者 | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2002-06-05 | | 公開日 | 2003-07-29 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of MTH1187 and its Yeast Ortholog YBL001C
Proteins, 52, 2003
|
|
4I3F
 
 | | Crystal structure of serine hydrolase CCSP0084 from the polyaromatic hydrocarbon (PAH)-degrading bacterium Cycloclasticus zankles | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | 著者 | Stogios, P.J, Xu, X, Dong, A, Cui, H, Alcaide, M, Tornes, J, Gertler, C, Yakimov, M.M, Golyshin, P.N, Ferrer, M, Savchenko, A. | | 登録日 | 2012-11-26 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Single residues dictate the co-evolution of dual esterases: MCP hydrolases from the alpha / beta hydrolase family.
Biochem.J., 454, 2013
|
|
