5GN6
 
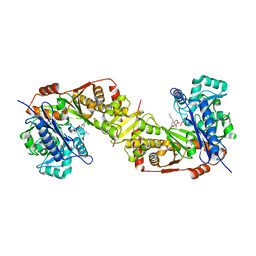 | | Crystal structure of glycerol kinase from Trypanosoma brucei gambiense complexed with cumarin derivative-17b | | 分子名称: | 4-butyl-7,8-bis(oxidanyl)chromen-2-one, GLYCEROL, Glycerol kinase | | 著者 | Balogun, E.O, Inaoka, D.K, Shiba, T, Tsuge, T, May, B, Sato, T, Kido, Y, Takeshi, N, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Michels, P.A.M, Watanabe, Y, Moore, A.L, Harada, S, Kita, K. | | 登録日 | 2016-07-19 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery of trypanocidal coumarins with dual inhibition of both the glycerol kinase and alternative oxidase ofTrypanosoma brucei brucei.
Faseb J., 33, 2019
|
|
2SIC
 
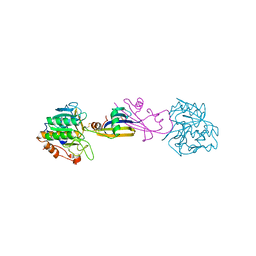 | | REFINED CRYSTAL STRUCTURE OF THE COMPLEX OF SUBTILISIN BPN' AND STREPTOMYCES SUBTILISIN INHIBITOR AT 1.8 ANGSTROMS RESOLUTION | | 分子名称: | CALCIUM ION, STREPTOMYCES SUBTILISIN INHIBITOR (SSI), SUBTILISIN BPN' | | 著者 | Mitsui, Y, Takeuchi, Y, Hirono, S, Akagawa, H, Nakamura, K.T. | | 登録日 | 1991-04-01 | | 公開日 | 1993-04-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Refined crystal structure of the complex of subtilisin BPN' and Streptomyces subtilisin inhibitor at 1.8 A resolution.
J.Mol.Biol., 221, 1991
|
|
7BQ3
 
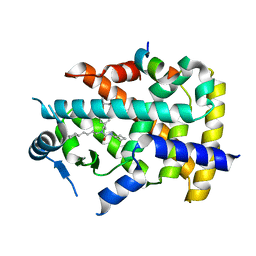 | | X-ray structure of human PPARalpha ligand binding domain-GW7647-SRC1 coactivator peptide co-crystals obtained by delipidation and co-crystallization | | 分子名称: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | 登録日 | 2020-03-23 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7BQ4
 
 | | X-ray structure of human PPARalpha ligand binding domain-eicosapentaenoic acid (EPA)-SRC1 coactivator peptide co-crystals obtained by delipidation and co-crystallization | | 分子名称: | 15-meric peptide from Nuclear receptor coactivator 1, 5,8,11,14,17-EICOSAPENTAENOIC ACID, GLYCEROL, ... | | 著者 | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | 登録日 | 2020-03-23 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7BPZ
 
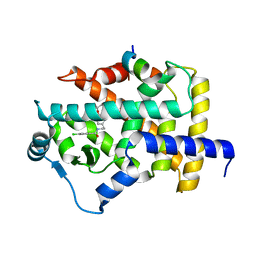 | | X-ray structure of human PPARalpha ligand binding domain-bezafibrate-SRC1 coactivator peptide co-crystals obtained by soaking | | 分子名称: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[P-[2-P-CHLOROBENZAMIDO)ETHYL]PHENOXY]-2-METHYLPROPIONIC ACID, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | 登録日 | 2020-03-23 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7BQ0
 
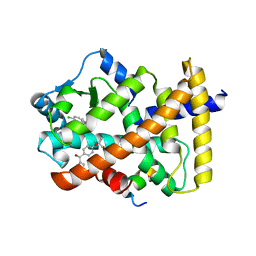 | | X-ray structure of human PPARalpha ligand binding domain-fenofibric acid-SRC1 coactivator peptide co-crystals obtained by delipidation and co-crystallization | | 分子名称: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | 登録日 | 2020-03-23 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.771 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7BQ1
 
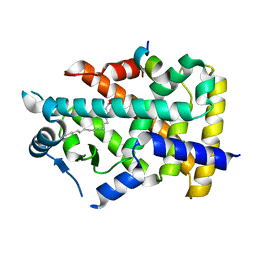 | | X-ray structure of human PPARalpha ligand binding domain-intrinsic fatty acid (E. coli origin)-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | 分子名称: | 15-meric peptide from Nuclear receptor coactivator 1, GLYCEROL, PALMITIC ACID, ... | | 著者 | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | 登録日 | 2020-03-23 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.521 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7BPY
 
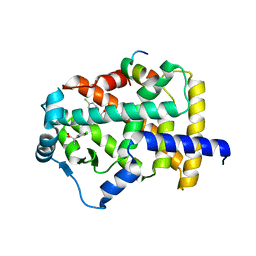 | | X-ray structure of human PPARalpha ligand binding domain-clofibric acid-SRC1 coactivator peptide co-crystals obtained by delipidation and co-crystallization | | 分子名称: | 15-meric peptide from Nuclear receptor coactivator 1, 2-(4-chloranylphenoxy)-2-methyl-propanoic acid, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | 登録日 | 2020-03-23 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
7BQ2
 
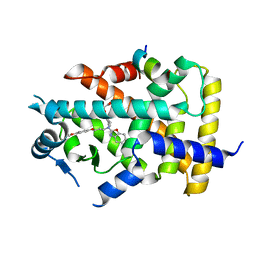 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate-SRC1 coactivator peptide co-crystals obtained by soaking | | 分子名称: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, 15-meric peptide from Nuclear receptor coactivator 1, GLYCEROL, ... | | 著者 | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | 登録日 | 2020-03-23 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6OZC
 
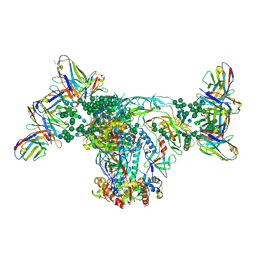 | | BG505 SOSIP.664 with 2G12 Fab2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2G12 Fab Heavy chain, ... | | 著者 | Cottrell, C.A, Ward, A.B. | | 登録日 | 2019-05-15 | | 公開日 | 2020-05-20 | | 最終更新日 | 2020-08-19 | | 実験手法 | ELECTRON MICROSCOPY (3.79 Å) | | 主引用文献 | Networks of HIV-1 Envelope Glycans Maintain Antibody Epitopes in the Face of Glycan Additions and Deletions.
Structure, 28, 2020
|
|
5JH9
 
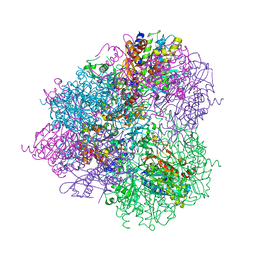 | | Crystal structure of prApe1 | | 分子名称: | CACODYLATE ION, Vacuolar aminopeptidase 1, ZINC ION | | 著者 | Noda, N.N, Adachi, W, Inagaki, F. | | 登録日 | 2016-04-20 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
5JGF
 
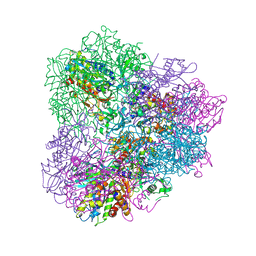 | | Crystal structure of mApe1 | | 分子名称: | Vacuolar aminopeptidase 1, ZINC ION | | 著者 | Noda, N.N, Adachi, W, Inagaki, F. | | 登録日 | 2016-04-20 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
5JHC
 
 | |
7SGF
 
 | | Lassa virus glycoprotein construct (Josiah GPC-I53-50A) in complex with LAVA01 antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Antanasijevic, A, Brouwer, P.J.M, Ward, A.B. | | 登録日 | 2021-10-05 | | 公開日 | 2022-10-12 | | 最終更新日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (4.41 Å) | | 主引用文献 | Lassa virus glycoprotein nanoparticles elicit neutralizing antibody responses and protection.
Cell Host Microbe, 30, 2022
|
|
7SGE
 
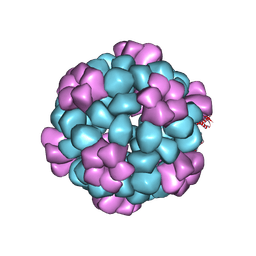 | |
7SGD
 
 | | Lassa virus glycoprotein construct(Josiah GPCysR4) recovered from GPC-I53-50 nanoparticle by localized reconstruction | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Antanasijevic, A, Brouwer, P.J.M, Ward, A.B. | | 登録日 | 2021-10-05 | | 公開日 | 2022-10-12 | | 最終更新日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (3.97 Å) | | 主引用文献 | Lassa virus glycoprotein nanoparticles elicit neutralizing antibody responses and protection.
Cell Host Microbe, 30, 2022
|
|
5ETY
 
 | | Crystal Structure of human Tankyrase-1 bound to K-756 | | 分子名称: | 3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-1,4-dihydroquinazolin-2-one, Tankyrase-1, ZINC ION | | 著者 | Takahashi, Y, Miyagi, H, Suzuki, M, Saito, J. | | 登録日 | 2015-11-18 | | 公開日 | 2016-06-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The Discovery and Characterization of K-756, a Novel Wnt/ beta-Catenin Pathway Inhibitor Targeting Tankyrase
Mol.Cancer Ther., 15, 2016
|
|
5EVY
 
 | | Salicylate hydroxylase substrate complex | | 分子名称: | 2-HYDROXYBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Salicylate hydroxylase | | 著者 | Morimoto, Y, Uemura, T. | | 登録日 | 2015-11-20 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | The catalytic mechanism of decarboxylative hydroxylation of salicylate hydroxylase revealed by crystal structure analysis at 2.5 angstrom resolution
Biochem.Biophys.Res.Commun., 469, 2016
|
|
5WQJ
 
 | |
5WQK
 
 | |
3SIC
 
 | |
8J6N
 
 | | Crystal structure of Cystathionine gamma-lyase in complex with compound 1 | | 分子名称: | 1,2-ETHANEDIOL, Cystathionine gamma-lyase, GLYCEROL, ... | | 著者 | Hibi, R, Toma-Fukai, S, Shimizu, T, Hanaoka, K. | | 登録日 | 2023-04-26 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of a cystathionine gamma-lyase (CSE) selective inhibitor targeting active-site pyridoxal 5'-phosphate (PLP) via Schiff base formation.
Sci Rep, 13, 2023
|
|
1WQV
 
 | | Human Factor Viia-Tissue Factor Complexed with propylsulfonamide-D-Thr-Met-p-aminobenzamidine | | 分子名称: | CALCIUM ION, Coagulation factor VII, N-[DIHYDROXY(PROPYL)-LAMBDA~4~-SULFANYL]THREONYL-N~1~-{4-[AMINO(IMINO)METHYL]BENZYL}METHIONINAMIDE, ... | | 著者 | Kadono, S, Sakamoto, A, Kikuchi, Y, Oh-eda, M, Yabuta, N, Koga, T, Hattori, K, Shiraishi, T, Haramura, M, Kodama, H. | | 登録日 | 2004-10-02 | | 公開日 | 2005-10-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of human factor VIIa/tissue factor in complex with peptide mimetic inhibitor
Biochem.Biophys.Res.Commun., 324, 2004
|
|
5E6O
 
 | | Crystal structure of C. elegans LGG-2 bound to an AIM/LIR motif | | 分子名称: | Protein lgg-2, TRP-GLU-GLU-LEU | | 著者 | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | 登録日 | 2015-10-10 | | 公開日 | 2016-01-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
5E6N
 
 | | Crystal structure of C. elegans LGG-2 | | 分子名称: | Protein lgg-2 | | 著者 | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | 登録日 | 2015-10-10 | | 公開日 | 2016-01-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
