3S0B
 
 | | Apis mellifera OBP14 in complex with the fluorescent probe 1-N-phenylnaphthylamine (NPN) | | 分子名称: | N-phenylnaphthalen-1-amine, OBP14 | | 著者 | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | 登録日 | 2011-05-13 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3S0A
 
 | | Apis mellifera OBP14, native apo-protein | | 分子名称: | OBP14 | | 著者 | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | 登録日 | 2011-05-13 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3S0G
 
 | | Apis mellifera OBP 14 double mutant Gln44Cys, His97Cys | | 分子名称: | OBP14 | | 著者 | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | 登録日 | 2011-05-13 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3RZS
 
 | | Apis mellifera OBP14 in complex with Ta6Br14 | | 分子名称: | HEXATANTALUM DODECABROMIDE, OBP14 | | 著者 | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | 登録日 | 2011-05-12 | | 公開日 | 2011-11-30 | | 最終更新日 | 2012-01-11 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3S0D
 
 | | Apis mellifera OBP 14 in complex with the citrus odorant citralva (3,7-dimethylocta-2,6-dienenitrile) | | 分子名称: | (2Z)-3,7-dimethylocta-2,6-dienenitrile, OBP14 | | 著者 | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | 登録日 | 2011-05-13 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3S0E
 
 | | Apis mellifera OBP14 in complex with the odorant eugenol (2-methoxy-4(2-propenyl)-phenol) | | 分子名称: | 2-methoxy-4-(prop-2-en-1-yl)phenol, OBP14 | | 著者 | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | 登録日 | 2011-05-13 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
3S0F
 
 | | Apis mellifera OBP14 native apo, crystal form 2 | | 分子名称: | OBP14 | | 著者 | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | 登録日 | 2011-05-13 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
7PU5
 
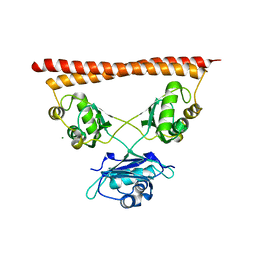 | | Structure of SFPQ-NONO complex | | 分子名称: | MAGNESIUM ION, Non-POU domain-containing octamer-binding protein, Splicing factor, ... | | 著者 | Fribourg, S. | | 登録日 | 2021-09-28 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.999 Å) | | 主引用文献 | Crystal structure of SFPQ-NONO heterodimer.
Biochimie, 198, 2022
|
|
2JLT
 
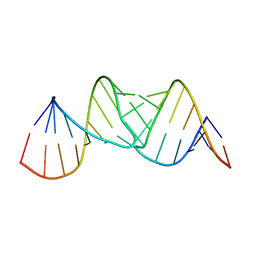 | |
4A97
 
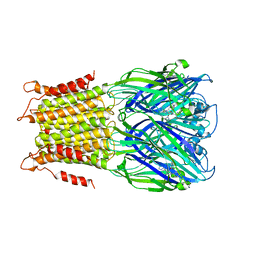 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with zopiclone | | 分子名称: | (5R)-6-(5-chloropyridin-2-yl)-7-oxo-6,7-dihydro-5H-pyrrolo[3,4-b]pyrazin-5-yl 4-methylpiperazine-1-carboxylate, CYS-LOOP LIGAND-GATED ION CHANNEL | | 著者 | Spurny, R, Brams, M, Ulens, C. | | 登録日 | 2011-11-24 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.343 Å) | | 主引用文献 | Pentameric Ligand-Gated Ion Channel Elic is Activated by Gaba and Modulated by Benzodiazepines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1J96
 
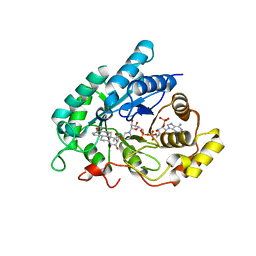 | | Human 3alpha-HSD type 3 in Ternary Complex with NADP and Testosterone | | 分子名称: | 3alpha-hydroxysteroid dehydrogenase type 3, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Nahoum, V, Labrie, F, Lin, S.-X. | | 登録日 | 2001-05-23 | | 公開日 | 2002-05-23 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Structure of the human 3alpha-hydroxysteroid dehydrogenase type 3 in complex with testosterone and NADP at 1.25-A resolution.
J.Biol.Chem., 276, 2001
|
|
7AV4
 
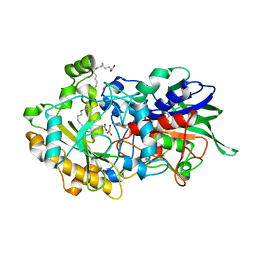 | | Dark state structure of the C432S mutant of Fatty Acid Photodecarboxylase (FAP) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | 著者 | Schlichting, I, Hartmann, E, Arnoux, P, Sorigue, D, Beisson, F. | | 登録日 | 2020-11-04 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.936 Å) | | 主引用文献 | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
7Z6O
 
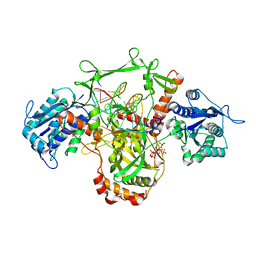 | | X-Ray studies of Ku70/80 reveal the binding site for IP6 | | 分子名称: | DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Varela, P.F, Charbonnier, J.B. | | 登録日 | 2022-03-14 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
7ZVT
 
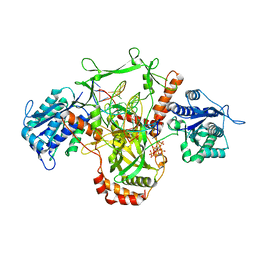 | | CryoEM structure of Ku heterodimer bound to DNA | | 分子名称: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | 登録日 | 2022-05-17 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
7ZT6
 
 | |
2X6S
 
 | | Human foamy virus integrase - catalytic core. Magnesium-bound structure. | | 分子名称: | INTEGRASE, MAGNESIUM ION | | 著者 | Rety, S, Delelis, O, Rezabkova, L, Dubanchet, B, Silhan, J, Lewit-Bentley, A. | | 登録日 | 2010-02-19 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural Studies of the Catalytic Core of the Primate Foamy Virus (Pfv-1) Integrase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2X6N
 
 | | Human foamy virus integrase - catalytic core. Manganese-bound structure. | | 分子名称: | INTEGRASE, MANGANESE (II) ION | | 著者 | Rety, S, Delelis, O, Rezabkova, L, Dubanchet, B, Silhan, J, Lewit-Bentley, A. | | 登録日 | 2010-02-18 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Structural Studies of the Catalytic Core of the Primate Foamy Virus (Pfv-1) Integrase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4NLC
 
 | |
6H6P
 
 | | UbiJ-SCP2 Ubiquinone synthesis protein | | 分子名称: | CALCIUM ION, PENTAETHYLENE GLYCOL, Ubiquinone biosynthesis protein UbiJ | | 著者 | Fyfe, C.D, Legrand, P, Pecqueur, L, Ciccone, L, Lombard, M, Fontecave, M. | | 登録日 | 2018-07-28 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A Soluble Metabolon Synthesizes the Isoprenoid Lipid Ubiquinone.
Cell Chem Biol, 26, 2019
|
|
6Z74
 
 | |
7Q5A
 
 | | Lanreotide nanotube | | 分子名称: | Lanreotide | | 著者 | Pieri, L, Wang, F, Arteni, A.A, Bressanelli, S, Egelman, E.H, Paternostre, M. | | 登録日 | 2021-11-03 | | 公開日 | 2022-04-06 | | 実験手法 | ELECTRON MICROSCOPY (2.46 Å) | | 主引用文献 | Atomic structure of Lanreotide nanotubes revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8CH1
 
 | | PBP AccA from A. vitis S4 in complex with Agrocinopine A | | 分子名称: | 1,2-ETHANEDIOL, 2-O-phosphono-alpha-L-arabinopyranose, 2-O-phosphono-beta-L-arabinopyranose, ... | | 著者 | Morera, S, Vigouroux, A. | | 登録日 | 2023-02-06 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.496 Å) | | 主引用文献 | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CKO
 
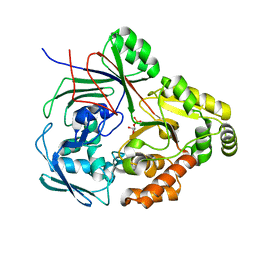 | | PBP AccA from A.tumefaciens C58 in complex with agrocinopine C-like | | 分子名称: | 2-O-phosphono-alpha-D-glucopyranose, 2-O-phosphono-beta-D-glucopyranose, ABC transporter substrate-binding protein, ... | | 著者 | Morera, S, Vigouroux, A. | | 登録日 | 2023-02-16 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.421 Å) | | 主引用文献 | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CB9
 
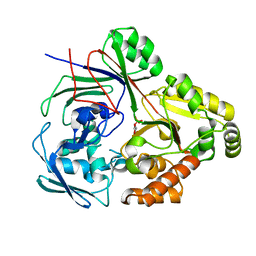 | | PBP AccA from A. tumefaciens Bo542 in complex with D-Glucose-2-phosphate | | 分子名称: | 2-O-phosphono-alpha-D-glucopyranose, 2-O-phosphono-beta-D-glucopyranose, Agrocinopine utilization periplasmic binding protein AccA | | 著者 | Morera, S, Vigouroux, A. | | 登録日 | 2023-01-25 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CAY
 
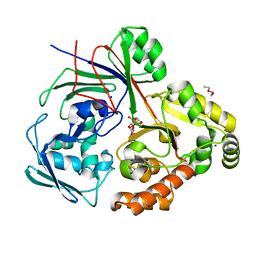 | | PBP AccA from A. tumefaciens Bo542 in complex with Agrocinopine D-like | | 分子名称: | Agrocinopine D-like (C2-C2 linked; with an alpha and beta-D-glucopyranose), Agrocinopine D-like (C2-C2 linked; with two alpha-D-glucopyranoses), Agrocinopine utilization periplasmic binding protein AccA, ... | | 著者 | Morera, S, Vigouroux, A, Siragu, S. | | 登録日 | 2023-01-24 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.626 Å) | | 主引用文献 | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
