8IPQ
 
 | |
6UH3
 
 | | Crystal structure of bacterial heliorhodopsin 48C12 | | 分子名称: | DI(HYDROXYETHYL)ETHER, Heliorhodopsin, PALMITIC ACID, ... | | 著者 | Lu, Y, Zhou, X.E, Gao, X, Xia, R, Xu, Z, Wang, N, Leng, Y, Melcher, K, Xu, H.E, He, Y. | | 登録日 | 2019-09-26 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of heliorhodopsin 48C12.
Cell Res., 30, 2020
|
|
7D1M
 
 | | CRYSTAL STRUCTURE OF THE SARS-CoV-2 MAIN PROTEASE COMPLEXED WITH GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fu, L.F, Gilski, M, Shabalin, I, Gao, G.F, Qi, J.X. | | 登録日 | 2020-09-14 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease.
Nat Commun, 11, 2020
|
|
6L10
 
 | | PHF20L1 Tudor1 - MES | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PHD finger protein 20-like protein 1, SULFATE ION | | 著者 | Lv, M.Q, Gao, J. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Conformational Selection in Ligand Recognition by the First Tudor Domain of PHF20L1.
J Phys Chem Lett, 11, 2020
|
|
6L1F
 
 | |
8IPI
 
 | |
8IPK
 
 | |
8IPL
 
 | |
8IPM
 
 | |
7RTC
 
 | |
4DDP
 
 | |
6L3F
 
 | |
6L8K
 
 | | Structure of URT1 in complex with UTP | | 分子名称: | URIDINE 5'-TRIPHOSPHATE, UTP:RNA uridylyltransferase 1 | | 著者 | Lingru, Z. | | 登録日 | 2019-11-06 | | 公開日 | 2020-01-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.999 Å) | | 主引用文献 | Crystal structure of Arabidopsis terminal uridylyl transferase URT1.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
7DHX
 
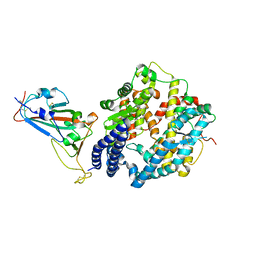 | | Crystal structure of SARS-CoV-2 RBD binding to pangolin ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ZINC ION, ... | | 著者 | Wang, Q.H, Qi, J.X, Wu, L.L. | | 登録日 | 2020-11-17 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular basis of pangolin ACE2 engaged by COVID-19 virus
Chin.Sci.Bull., 66, 2021
|
|
7KDT
 
 | |
8T60
 
 | |
3PXG
 
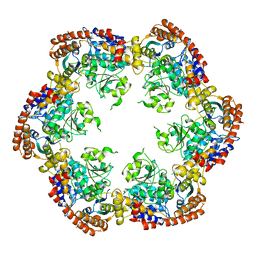 | | Structure of MecA121 and ClpC1-485 complex | | 分子名称: | Adapter protein mecA 1, Negative regulator of genetic competence ClpC/MecB | | 著者 | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | 登録日 | 2010-12-09 | | 公開日 | 2011-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.654 Å) | | 主引用文献 | Structure and mechanism of the hexameric MecA-ClpC molecular machine.
Nature, 471, 2011
|
|
3KDU
 
 | |
3KDT
 
 | |
6M6A
 
 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Shi, J, Wen, A, Feng, Y. | | 登録日 | 2020-03-14 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6M6C
 
 | | CryoEM structure of Thermus thermophilus RNA polymerase elongation complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Shi, J, Wen, A, Feng, Y. | | 登録日 | 2020-03-14 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
4RDO
 
 | | Structure of YTH-YTHDF2 in the free state | | 分子名称: | SULFATE ION, YTH domain-containing family protein 2 | | 著者 | Li, F.D, Zhao, D.B, Wu, J.H, Shi, Y.Y. | | 登録日 | 2014-09-19 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition.
Cell Res., 24, 2014
|
|
4RDN
 
 | | Structure of YTH-YTHDF2 in complex with m6A | | 分子名称: | N-methyladenosine, SULFATE ION, YTH domain-containing family protein 2 | | 著者 | Li, F.D, Zhao, D.B, Wu, J.H, Shi, Y.Y. | | 登録日 | 2014-09-19 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition.
Cell Res., 24, 2014
|
|
8TLM
 
 | | Structure of a class A GPCR/Fab complex | | 分子名称: | C-C chemokine receptor type 8, Green fluorescent protein fusion, Fab heavy chain, ... | | 著者 | Sun, D, Johnson, M, Masureel, M. | | 登録日 | 2023-07-27 | | 公開日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
8U1U
 
 | | Structure of a class A GPCR/agonist complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 1,C-C chemokine receptor type 8,EGFP fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Sun, D, Johnson, M, Masureel, M. | | 登録日 | 2023-09-02 | | 公開日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
