5UJ4
 
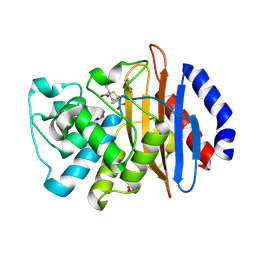 | | Crystal structure of the KPC-2 beta-lactamase complexed with hydrolyzed faropenem | | 分子名称: | (2R)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-5-[(2R)-oxolan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | 著者 | Pemberton, O.A, Chen, Y. | | 登録日 | 2017-01-16 | | 公開日 | 2017-04-26 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Molecular Basis of Substrate Recognition and Product Release by the Klebsiella pneumoniae Carbapenemase (KPC-2).
J. Med. Chem., 60, 2017
|
|
8I13
 
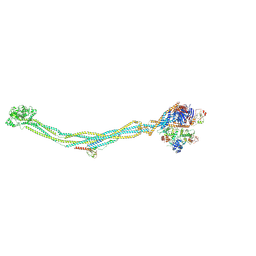 | | Cryo-EM structure of 6-subunit Smc5/6 | | 分子名称: | MMS21 isoform 1, NSE3 isoform 1, Non-structural maintenance of chromosomes element 1 homolog, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Duo, J, Zhenguo, C, Wang, L. | | 登録日 | 2023-01-12 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I4V
 
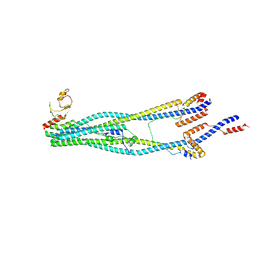 | | Cryo-EM structure of 5-subunit Smc5/6 arm region | | 分子名称: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Zhenguo, C, Wang, L. | | 登録日 | 2023-01-21 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (5.97 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I4W
 
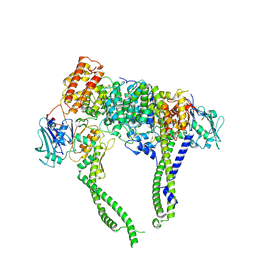 | | Cryo-EM structure of 5-subunit Smc5/6 head region | | 分子名称: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 5, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Cheng, T, Duo, J, Zhenguo, C, Wang, L. | | 登録日 | 2023-01-21 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (6.01 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I4X
 
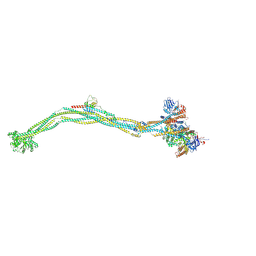 | | Cryo-EM structure of 5-subunit Smc5/6 | | 分子名称: | E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 5, Nse6, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | 登録日 | 2023-01-21 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (8.5 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I21
 
 | | Cryo-EM structure of 6-subunit Smc5/6 arm region | | 分子名称: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | 著者 | Jun, Z, Qian, L, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | 登録日 | 2023-01-13 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (6.02 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
7TOB
 
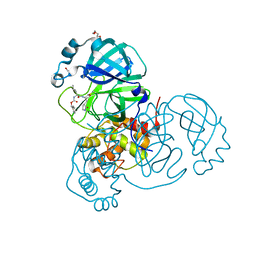 | | Crystal structure of the SARS-CoV-2 Omicron main protease (Mpro) in complex with inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | 著者 | Sacco, M.D, Wang, J, Chen, Y. | | 登録日 | 2022-01-24 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition.
Cell Res., 32, 2022
|
|
1T34
 
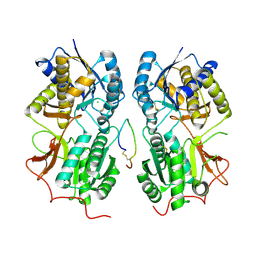 | | ROTATION MECHANISM FOR TRANSMEMBRANE SIGNALING BY THE ATRIAL NATRIURETIC PEPTIDE RECEPTOR | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Atrial natriuretic peptide factor, Atrial natriuretic peptide receptor A, ... | | 著者 | Ogawa, H, Qiu, Y, Ogata, C.M, Misono, K.S. | | 登録日 | 2004-04-23 | | 公開日 | 2004-08-03 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Crystal structure of hormone-bound atrial natriuretic peptide receptor extracellular domain: rotation mechanism for transmembrane signal transduction
J.Biol.Chem., 279, 2004
|
|
2L83
 
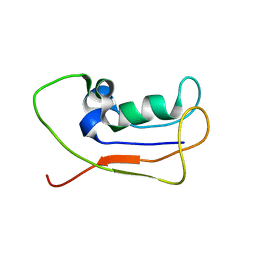 | | A protein from Haloferax volcanii | | 分子名称: | Small archaeal modifier protein 1 | | 著者 | Zhang, W, Liao, S, Fan, K, Tu, X. | | 登録日 | 2011-01-03 | | 公開日 | 2012-01-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Ionic strength-dependent conformations of a ubiquitin-like small archaeal modifier protein (SAMP1) from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
2IBJ
 
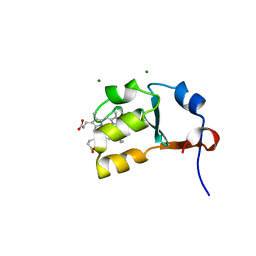 | | Structure of House Fly Cytochrome B5 | | 分子名称: | Cytochrome b5, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Terzyan, S, Zhang, X.C, Benson, D.R, Wang, L. | | 登録日 | 2006-09-11 | | 公開日 | 2007-02-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Comparison of cytochromes b(5) from insects and vertebrates.
Proteins, 67, 2007
|
|
8JEL
 
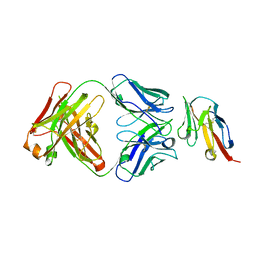 | | Crystal structure of TIGIT in complexed with Ociperlimab, crystal form I | | 分子名称: | T-cell immunoreceptor with Ig and ITIM domains, antibody heavy chain, antibody light chain | | 著者 | Sun, J, Zhang, X.X, Song, J. | | 登録日 | 2023-05-16 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEP
 
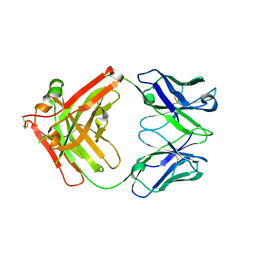 | | Crystal structure of Ociperlimab | | 分子名称: | antibody heavy chain, antibody light chain | | 著者 | Sun, J, Zhang, X.X, Song, J. | | 登録日 | 2023-05-16 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEQ
 
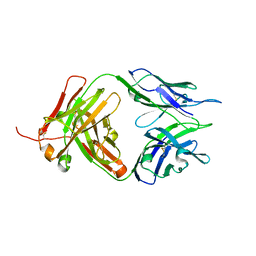 | | Crystal structure of Tiragolumab | | 分子名称: | antibody heavy chain, antibody light chain | | 著者 | Sun, J, Zhang, X.X, Song, J. | | 登録日 | 2023-05-16 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEN
 
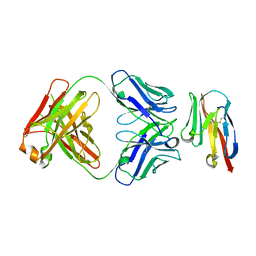 | | Crystal structure of TIGIT in complexed with Ociperlimab, crystal form II | | 分子名称: | T-cell immunoreceptor with Ig and ITIM domains, antibody heavy chain, antibody light chain | | 著者 | Sun, J, Zhang, X.X, Song, J. | | 登録日 | 2023-05-16 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEO
 
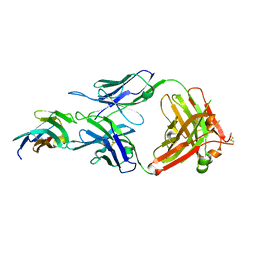 | |
8W8D
 
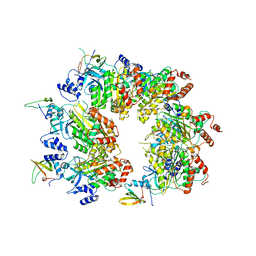 | |
8XH7
 
 | |
8XH6
 
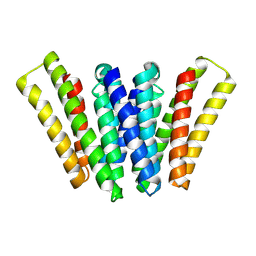 | | Structure of EBV LMP1 dimer | | 分子名称: | Latent membrane protein 1 | | 著者 | Gao, P, Huang, J.F. | | 登録日 | 2023-12-17 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Assembly and activation of EBV latent membrane protein 1.
Cell, 2024
|
|
1RA4
 
 | |
7LNI
 
 | | SeMet CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | 著者 | Horton, J.R, Cheng, X, Zhou, J. | | 登録日 | 2021-02-07 | | 公開日 | 2021-05-19 | | 最終更新日 | 2021-07-14 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Clostridioides difficile specific DNA adenine methyltransferase CamA squeezes and flips adenine out of DNA helix.
Nat Commun, 12, 2021
|
|
7LT5
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Cofactor SAH | | 分子名称: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | 著者 | Horton, J.R, Cheng, X, Zhou, J. | | 登録日 | 2021-02-18 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Clostridioides difficile specific DNA adenine methyltransferase CamA squeezes and flips adenine out of DNA helix.
Nat Commun, 12, 2021
|
|
7LNJ
 
 | |
8HQS
 
 | | Cryo-EM structure of 8-subunit Smc5/6 head region | | 分子名称: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, ... | | 著者 | Qian, L, Jun, Z, Xiang, Z, Tong, C, Wang, Z, Duo, J, Zhenguo, C, Wang, L. | | 登録日 | 2022-12-14 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
7T60
 
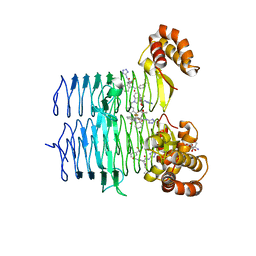 | | P. aeruginosa LpxA in complex with ligand L13 | | 分子名称: | (3S)-3-(5,5-dimethyl-2-oxo-1,3-oxazolidin-3-yl)-N-(1H-tetrazol-5-yl)-1-[3-(trifluoromethyl)benzoyl]-2,3-dihydro-1H-indole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, ... | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2021-12-13 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
7T61
 
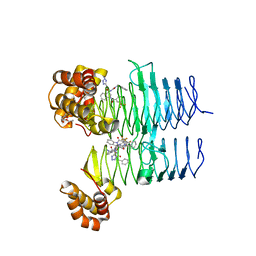 | | P. aeruginosa LpxA in complex with ligand L15 | | 分子名称: | (3S)-3-({[(Z)-phenylmethylidene]carbamoyl}amino)-N-(1H-tetrazol-5-yl)-1-[3-(trifluoromethyl)benzoyl]-2,3-dihydro-1H-indole-3-carboxamide, ACETATE ION, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, ... | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2021-12-13 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Based Ligand Design Targeting Pseudomonas aeruginosa LpxA in Lipid A Biosynthesis.
Acs Infect Dis., 8, 2022
|
|
