5K5L
 
 | |
5K5H
 
 | |
5KAE
 
 | |
5KE8
 
 | |
4X0N
 
 | |
4Q2K
 
 | | Bovine alpha chymotrypsin bound to a cyclic peptide inhibitor, 5b | | 分子名称: | (11S)-4,9-dioxo-N-[(2S)-1-oxo-3-phenylpropan-2-yl]-17,22-dioxa-10,30-diazatetracyclo[21.2.2.2~13,16~.1~5,8~]triaconta-1(25),5,7,13,15,23,26,28-octaene-11-carboxamide, Chymotrypsinogen A | | 著者 | Chan, H.Y, Bruning, J.B, Abell, A.D. | | 登録日 | 2014-04-09 | | 公開日 | 2014-07-23 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Macrocyclic protease inhibitors with reduced peptide character.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6LKQ
 
 | | The Structural Basis for Inhibition of Ribosomal Translocation by Viomycin | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Zhang, L, Wang, Y.H, Lancaster, L, Zhou, J, Noller, H.F. | | 登録日 | 2019-12-20 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The structural basis for inhibition of ribosomal translocation by viomycin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6M4S
 
 | | Crystal Structure Analysis of the cytochrome P450 CYP-Sb21 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Cytochrome P450 hydroxylase sb21, ... | | 著者 | Li, F.W, Li, S.Y. | | 登録日 | 2020-03-09 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-guided manipulation of the regioselectivity of the cyclosporine A hydroxylase CYP-sb21 from Sebekia benihana .
Synth Syst Biotechnol, 5, 2020
|
|
5K5J
 
 | |
5EDS
 
 | | Crystal structure of human PI3K-gamma in complex with benzimidazole inhibitor 5 | | 分子名称: | 4-azanyl-6-[[(1~{S})-1-[6-fluoranyl-1-(3-methylsulfonylphenyl)benzimidazol-2-yl]ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | 著者 | Whittington, D.A, Tang, J, Yakowec, P. | | 登録日 | 2015-10-21 | | 公開日 | 2015-12-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery, Optimization, and in Vivo Evaluation of Benzimidazole Derivatives AM-8508 and AM-9635 as Potent and Selective PI3K delta Inhibitors.
J.Med.Chem., 59, 2016
|
|
5KE9
 
 | |
6UKG
 
 | |
5KEA
 
 | |
6UKF
 
 | |
6MC1
 
 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((9-(methylthio)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3,3-dimethyl-1-{[9-(methylsulfanyl)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl]sulfanyl}butan-2-one, ACETATE ION, ... | | 著者 | Gannam, Z.T.K, Anderson, K.S, Bennett, A.M, Lolis, E. | | 登録日 | 2018-08-30 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | An allosteric site on MKP5 reveals a strategy for small-molecule inhibition.
Sci.Signal., 13, 2020
|
|
2L7E
 
 | |
5KL6
 
 | | Wilms Tumor Protein (WT1) Q369R ZnF2-4 in complex with DNA | | 分子名称: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*GP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*CP*CP*CP*CP*AP*CP*GP*C)-3'), Wilms tumor protein, ... | | 著者 | Hashimoto, H, Cheng, X. | | 登録日 | 2016-06-23 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.641 Å) | | 主引用文献 | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
6UKI
 
 | |
6LRM
 
 | | Crystal structure of PDE4D catalytic domain in complex with arctigenin | | 分子名称: | 1,2-ETHANEDIOL, Arctigenin, MAGNESIUM ION, ... | | 著者 | Zhang, X.L, Li, M.J, Xu, Y.C. | | 登録日 | 2020-01-16 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Identification of phosphodiesterase-4 as the therapeutic target of arctigenin in alleviating psoriatic skin inflammation.
J Adv Res, 33, 2021
|
|
8SSU
 
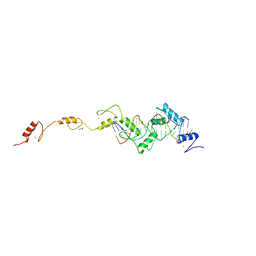 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 19mer DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (19-MER) Strand I, DNA (19-MER) Strand II, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SST
 
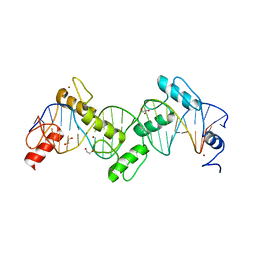 | | ZnFs 1-7 of CCCTC-binding factor (CTCF) K365T Mutant Complexed with 23mer | | 分子名称: | 1,2-ETHANEDIOL, DNA Strand (23mer) I, DNA Strand (23mer) II, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSS
 
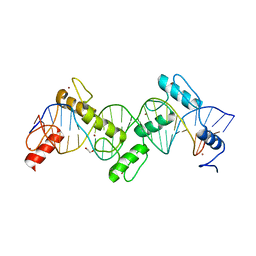 | | ZnFs 1-7 of CCCTC-binding factor (CTCF) Complexed with 23mer | | 分子名称: | 1,2-ETHANEDIOL, DNA Strand (23mer) I, DNA Strand (23mer) II, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSQ
 
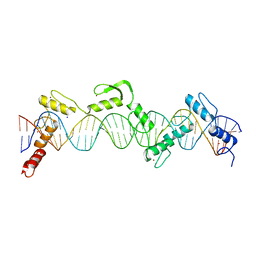 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-4 | | 分子名称: | DNA (35-MER) Strand 2, DNA (35-MER) Strand I, SODIUM ION, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.12 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
8SSR
 
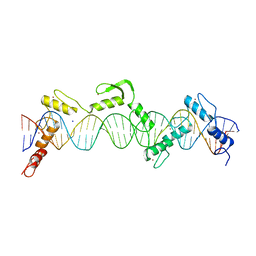 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 35mer DNA 35-20 | | 分子名称: | DNA (35-MER) Strand I, DNA (35-MER) Strand II, SODIUM ION, ... | | 著者 | Horton, J.R, Yang, J, Cheng, X. | | 登録日 | 2023-05-08 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
5KL3
 
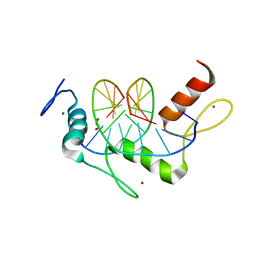 | | Wilms Tumor Protein (WT1) ZnF2-4 Q369H in complex with DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | 著者 | Hashimoto, H, Cheng, X. | | 登録日 | 2016-06-23 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.449 Å) | | 主引用文献 | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
