7E8K
 
 | |
7E8J
 
 | |
5GWE
 
 | | cytochrome P450 CREJ | | 分子名称: | (4-methylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Dong, S, liu, X, Wang, X, Feng, Y. | | 登録日 | 2016-09-11 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Selective oxidation of aliphatic C-H bonds in alkylphenols by a chemomimetic biocatalytic system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4Y4R
 
 | | Crystal structure of ribosomal oxygenase NO66 dimer mutant | | 分子名称: | ACETATE ION, Bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66, NICKEL (II) ION | | 著者 | Wang, C, Hang, T, Zang, J. | | 登録日 | 2015-02-11 | | 公開日 | 2015-10-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure of the JmjC domain-containing protein NO66 complexed with ribosomal protein Rpl8.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YR6
 
 | | Fab fragment of 5G6 in complex with epitope peptide | | 分子名称: | ACE-LYS-LEU-ARG-GLY-VAL-LEU-GLN-GLY-HIS-LEU, GLYCEROL, heavy chain of 5G6, ... | | 著者 | Tao, Y, Mo, X, Li, R. | | 登録日 | 2015-03-14 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Structural basis for the specific inhibition of glycoprotein Ib alpha shedding by an inhibitory antibody.
Sci Rep, 6, 2016
|
|
5IVY
 
 | |
5OLR
 
 | | Rhamnogalacturonan lyase | | 分子名称: | CALCIUM ION, PHOSPHATE ION, Rhamnogalacturonan lyase, ... | | 著者 | Basle, A, Luis, A.S, Gilbert, H.J. | | 登録日 | 2017-07-28 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides.
Nat Microbiol, 3, 2018
|
|
7TBI
 
 | | Composite structure of the S. cerevisiae nuclear pore complex (NPC) | | 分子名称: | Dyn2, Nic96 R1, Nic96 R2, ... | | 著者 | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-15 | | 最終更新日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (25 Å) | | 主引用文献 | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
5IW0
 
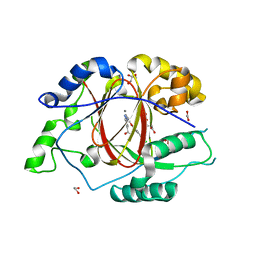 | |
7BZT
 
 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | 著者 | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | 登録日 | 2020-04-28 | | 公開日 | 2020-07-22 | | 最終更新日 | 2020-08-19 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BR8
 
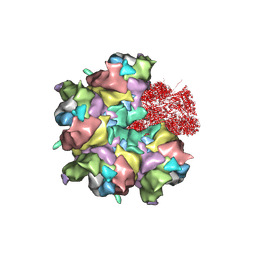 | | Epstein-Barr virus, C5 penton vertex, CATC absent. | | 分子名称: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | 著者 | Li, Z, Yu, X. | | 登録日 | 2020-03-26 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
7BZN
 
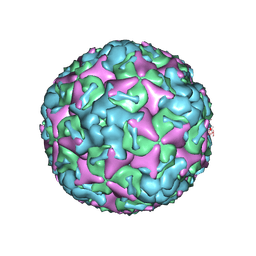 | | Cryo-EM structure of mature Coxsackievirus A10 at pH 7.4 | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | 登録日 | 2020-04-28 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BSI
 
 | |
7BQT
 
 | |
7WRI
 
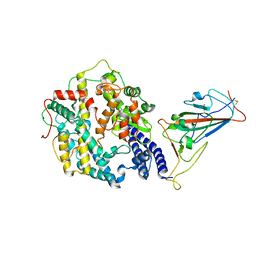 | | Cryo-EM structure of SARS-CoV-2 Omicron spike receptor-binding domain in complex with mouse ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein, ... | | 著者 | Han, P, Xie, Y, Qi, J. | | 登録日 | 2022-01-26 | | 公開日 | 2022-06-08 | | 最終更新日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
7WSK
 
 | | Crystal structure of SARS-CoV-2 Omicron spike receptor-binding domain in complex with civet ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | 著者 | Huang, B, Han, P, Qi, J. | | 登録日 | 2022-01-29 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
7BQX
 
 | | Epstein-Barr virus, C5 portal vertex | | 分子名称: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | 著者 | Li, Z, Yu, X. | | 登録日 | 2020-03-25 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
7BZO
 
 | | Cryo-EM structure of mature Coxsackievirus A10 at pH 5.5 | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | 登録日 | 2020-04-28 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BR7
 
 | | Epstein-Barr virus, C1 portal-proximal penton vertex, CATC binding | | 分子名称: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | 著者 | Li, Z, Yu, X. | | 登録日 | 2020-03-26 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
7BZU
 
 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | 著者 | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | 登録日 | 2020-04-28 | | 公開日 | 2020-07-22 | | 最終更新日 | 2020-08-19 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1F6W
 
 | |
5ISL
 
 | |
5OPJ
 
 | | Beta-L-arabinofuranosidase | | 分子名称: | Rhamnogalacturonan lyase, ZINC ION, alpha-L-arabinofuranose | | 著者 | Basle, A, Luis, A.S, Gilbert, H.J. | | 登録日 | 2017-08-10 | | 公開日 | 2018-02-28 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides.
Nat Microbiol, 3, 2018
|
|
5IVJ
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N11 [3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid] | | 分子名称: | 3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2016-03-21 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.569 Å) | | 主引用文献 | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
4ZS3
 
 | | Structural complex of 5-aminofluorescein bound to the FTO protein | | 分子名称: | 2-OXOGLUTARIC ACID, 5-amino-2-(6-hydroxy-3-oxo-3H-xanthen-9-yl)benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, ... | | 著者 | Wang, T, Hong, T, Huang, Y, Yang, C.G, Zhou, X. | | 登録日 | 2015-05-13 | | 公開日 | 2015-11-04 | | 最終更新日 | 2015-11-18 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Fluorescein Derivatives as Bifunctional Molecules for the Simultaneous Inhibiting and Labeling of FTO Protein
J.Am.Chem.Soc., 137, 2015
|
|
