2NV3
 
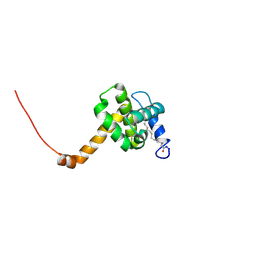 | | Solution structure of L8A mutant of HIV-1 myristoylated matrix protein | | 分子名称: | Gag polyprotein, MYRISTIC ACID | | 著者 | Saad, J.S, Loeliger, E, Luncsford, P, Liriano, M, Tai, J, Kim, A, Miller, J, Joshi, A, Freed, E.O, Summers, M.F. | | 登録日 | 2006-11-10 | | 公開日 | 2007-02-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Point mutations in the HIV-1 matrix protein turn off the myristyl switch.
J.Mol.Biol., 366, 2007
|
|
2O19
 
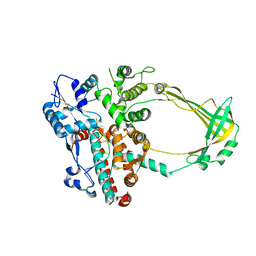 | | Structure of E. coli topoisomersae III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol at pH 5.5 | | 分子名称: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | 著者 | Changela, A, DiGate, R, Mondragon, A. | | 登録日 | 2006-11-28 | | 公開日 | 2007-04-03 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
7ALI
 
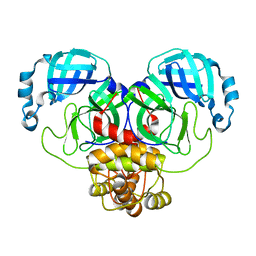 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup P2(1)). | | 分子名称: | 3C-like proteinase | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | 登録日 | 2020-10-06 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7UM0
 
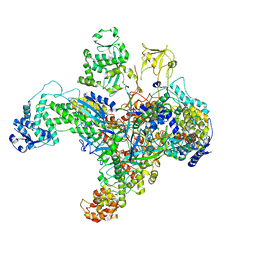 | | Structure of the phage AR9 non-virion RNA polymerase holoenzyme in complex with two DNA oligonucleotides containing the AR9 P077 promoter as determined by cryo-EM | | 分子名称: | DNA (5'-D(P*GP*UP*U)-3'), DNA-directed RNA polymerase, DNA-directed RNA polymerase beta subunit, ... | | 著者 | Leiman, P.G, Fraser, A, Sokolova, M.L. | | 登録日 | 2022-04-05 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of template strand deoxyuridine promoter recognition by a viral RNA polymerase.
Nat Commun, 13, 2022
|
|
6TPJ
 
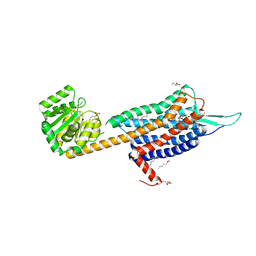 | | Crystal structure of the Orexin-2 receptor in complex with suvorexant at 2.76 A resolution | | 分子名称: | AMMONIUM ION, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Hypocretin receptor-2, ... | | 著者 | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | 登録日 | 2019-12-13 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6TQ7
 
 | | Crystal structure of the Orexin-1 receptor in complex with SB-334867 | | 分子名称: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 1-(2-methyl-1,3-benzoxazol-6-yl)-3-(1,5-naphthyridin-4-yl)urea, Orexin receptor type 1, ... | | 著者 | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | 登録日 | 2019-12-16 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.6636 Å) | | 主引用文献 | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
2O9X
 
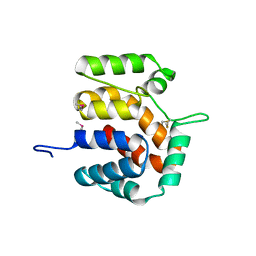 | | Crystal Structure Of A Putative Redox Enzyme Maturation Protein From Archaeoglobus Fulgidus | | 分子名称: | Reductase, assembly protein | | 著者 | Kirillova, O, Chruszcz, M, Skarina, T, Gorodichtchenskaia, E, Cymborowski, M, Shumilin, I, Savchenko, A, Edwards, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-12-14 | | 公開日 | 2007-01-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | An extremely SAD case: structure of a putative redox-enzyme maturation protein from Archaeoglobus fulgidus at 3.4 A resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
7UM1
 
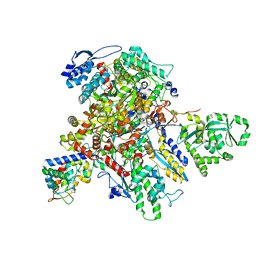 | | Structure of bacteriophage AR9 non-virion RNAP polymerase holoenzyme determined by cryo-EM | | 分子名称: | DNA-directed RNA polymerase, DNA-directed RNA polymerase beta subunit, DNA-directed RNA polymerase beta' subunit, ... | | 著者 | Leiman, P.G, Fraser, A, Sokolova, M.L. | | 登録日 | 2022-04-05 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural basis of template strand deoxyuridine promoter recognition by a viral RNA polymerase.
Nat Commun, 13, 2022
|
|
6VUY
 
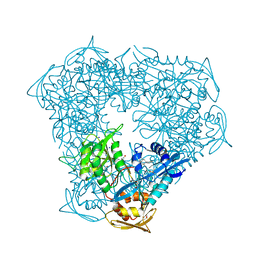 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT358 | | 分子名称: | (7S)-7-phenyl-2-{[3-(piperidin-1-yl)propyl]sulfanyl}-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4-amine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | 著者 | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | 登録日 | 2020-02-16 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
7V01
 
 | | Staphylococcus epidermidis RP62a CRISPR short effector complex with self RNA target and ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, ... | | 著者 | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | 登録日 | 2022-05-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7UZY
 
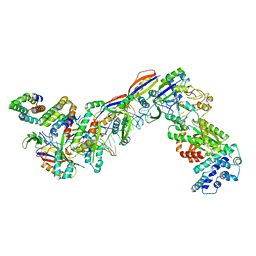 | | Staphylococcus epidermidis RP62A CRISPR effector complex with non-self target RNA 2 | | 分子名称: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | 著者 | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | 登録日 | 2022-05-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
6VPC
 
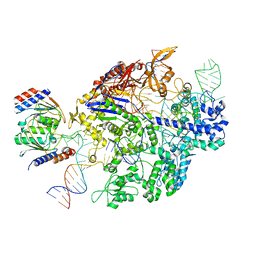 | | Structure of the SpCas9 DNA adenine base editor - ABE8e | | 分子名称: | CRISPR-associated endonuclease Cas9, Cas9 (SpCas9) single-guide RNA (sgRNA), DNA non-target strand (NTS), ... | | 著者 | Knott, G.J, Lapinaite, A, Doudna, J.A. | | 登録日 | 2020-02-03 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | DNA capture by a CRISPR-Cas9-guided adenine base editor.
Science, 369, 2020
|
|
7V02
 
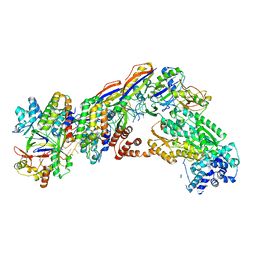 | | Staphylococcus epidermidis RP62A CRISPR short effector complex | | 分子名称: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | 著者 | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | 登録日 | 2022-05-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (4.97 Å) | | 主引用文献 | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
6TOT
 
 | | Crystal structure of the Orexin-1 receptor in complex with lemborexant | | 分子名称: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (1~{R},2~{S})-2-[(2,4-dimethylpyrimidin-5-yl)oxymethyl]-~{N}-(5-fluoranylpyridin-2-yl)-2-(3-fluorophenyl)cyclopropane-1-carboxamide, Orexin receptor type 1, ... | | 著者 | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | 登録日 | 2019-12-11 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
7UZZ
 
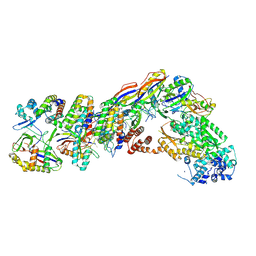 | | Staphylococcus epidermidis RP62a CRISPR tall effector complex | | 分子名称: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | 著者 | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | 登録日 | 2022-05-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (4.45 Å) | | 主引用文献 | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
2OGM
 
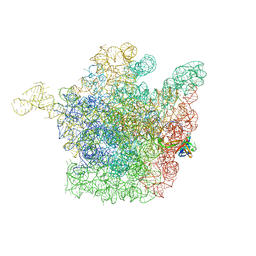 | | The crystal structure of the large ribosomal subunit from Deinococcus radiodurans complexed with the pleuromutilin derivative SB-571519 | | 分子名称: | (2S,3AR,4R,5S,6S,8R,9R,9AR,10R)-2,5-DIHYDROXY-4,6,9,10-TETRAMETHYL-1-OXO-6-VINYLDECAHYDRO-3A,9-PROP[1]ENOCYCLOPENTA[8]ANNULEN-8-YL [(6-AMINOPYRIDAZIN-3-YL)CARBONYL]CARBAMATE, 23S ribosomal RNA, 50S ribosomal protein L3 | | 著者 | Davidovich, C, Bashan, A, Auerbach-Nevo, T, Yonath, A. | | 登録日 | 2007-01-07 | | 公開日 | 2007-05-01 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6TP3
 
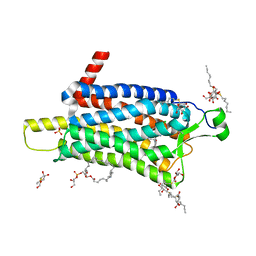 | | Crystal structure of the Orexin-1 receptor in complex with daridorexant | | 分子名称: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Orexin receptor type 1, SULFATE ION, ... | | 著者 | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | 登録日 | 2019-12-12 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.04 Å) | | 主引用文献 | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
7UZW
 
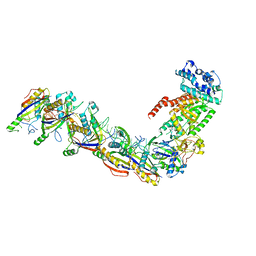 | | Staphylococcus epidermidis RP62a CRISPR effector subcomplex | | 分子名称: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm4, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | 著者 | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | 登録日 | 2022-05-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.55 Å) | | 主引用文献 | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7UZX
 
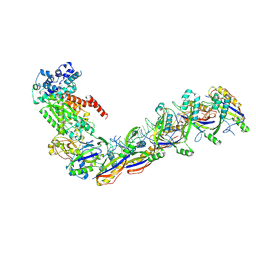 | | Staphylococcus epidermidis RP62a CRISPR effector subcomplex with non-self target RNA bound | | 分子名称: | CRISPR non-self RNA target, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm4, ... | | 著者 | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | 登録日 | 2022-05-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
6VZH
 
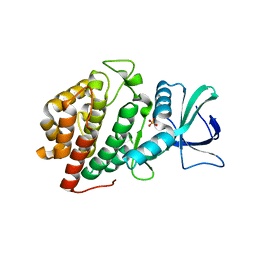 | | Structure of Human Vaccinia-related Kinase 1 (VRK1) Bound to LDSM311 | | 分子名称: | (7~{R})-2-[[3,5-bis(fluoranyl)-4-oxidanyl-phenyl]amino]-5,7-dimethyl-8-prop-2-ynyl-7~{H}-pteridin-6-one, 1,2-ETHANEDIOL, SULFATE ION, ... | | 著者 | dos Reis, C.V, Dutra, L.A, Gama, F, Ferreira, M, Mascarello, A, Azevedo, H, Guimaraes, C, Massirer, K.B, Arruda, P, Edwards, A.M, Counago, R.M, Structural Genomics Consortium (SGC) | | 登録日 | 2020-02-28 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structure of Human Vaccinia-related Kinase 1 (VRK1) Bound to LDSM311
To be Published
|
|
5NMW
 
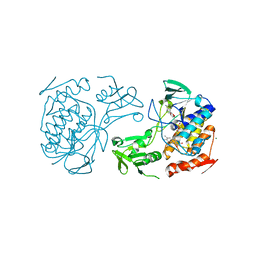 | | Crystal Structure of the pyrrolizidine alkaloid N-oxygenase from Zonocerus variegatus in complex with FAD | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, MAGNESIUM ION | | 著者 | Scheidig, A, Kubitza, C, Faust, A, Ober, D. | | 登録日 | 2017-04-07 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Crystal structure of pyrrolizidine alkaloid N-oxygenase from the grasshopper Zonocerus variegatus.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7V00
 
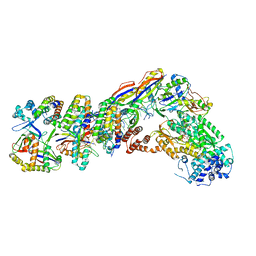 | | Staphylococcus epidermidis RP62a CRISPR tall effector complex with bound ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, ... | | 著者 | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | 登録日 | 2022-05-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
6W3R
 
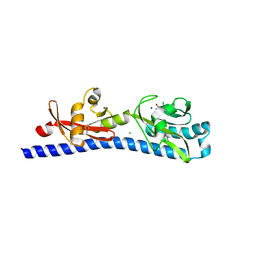 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with 3-methylisoleucine | | 分子名称: | 3-methyl-L-alloisoleucine, CHLORIDE ION, Methyl-accepting chemotaxis protein, ... | | 著者 | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | 登録日 | 2020-03-09 | | 公開日 | 2020-05-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
7AN5
 
 | | Enzyme of biosynthetic pathway | | 分子名称: | 3-[(1-Carboxyvinyl)oxy]benzoic acid, Chorismate dehydratase | | 著者 | Archna, A, Breithaupt, C, Stubbs, M.T. | | 登録日 | 2020-10-11 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis
J.Biol.Chem., 2022
|
|
6W4B
 
 | | The crystal structure of Nsp9 RNA binding protein of SARS CoV-2 | | 分子名称: | Non-structural protein 9 | | 著者 | Tan, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-10 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | The crystal structure of Nsp9 replicase protein of COVID-19
To Be Published
|
|
