7K65
 
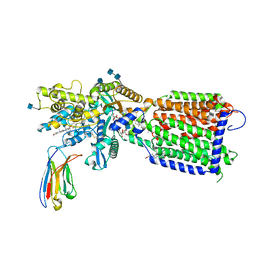 | | Hedgehog receptor Patched (PTCH1) in complex with conformation selective nanobody TI23 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | 著者 | Zhang, Y, Bulkley, D.P, Liang, J, Manglik, A, Cheng, Y, Beachy, P.A. | | 登録日 | 2020-09-18 | | 公開日 | 2021-03-17 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Hedgehog pathway activation through nanobody-mediated conformational blockade of the Patched sterol conduit.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7EEJ
 
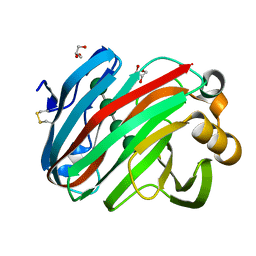 | | Complex structure of glycoside hydrolase family 12 beta-1,3-1,4-glucanase with cellobiose | | 分子名称: | GLYCEROL, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | 著者 | Jiang, Z.Q, Ma, J.W. | | 登録日 | 2021-03-18 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.47798049 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
7EE2
 
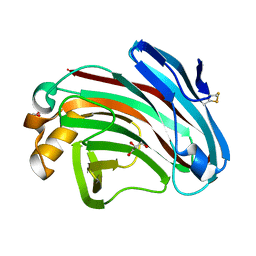 | | Structural insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.CQ31 | | 分子名称: | GLYCEROL, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | 著者 | Jiang, Z.Q, Ma, J. | | 登録日 | 2021-03-17 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.37011635 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
7EEE
 
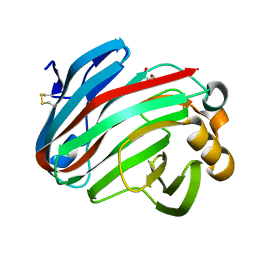 | | Complex structure of glycoside hydrolase family 12 beta-1,3-1,4-glucanase with gentiobiose | | 分子名称: | GLYCEROL, beta-D-mannopyranose-(1-6)-beta-D-mannopyranose, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | 著者 | Jiang, Z.Q, Ma, J.W. | | 登録日 | 2021-03-18 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.660792 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
6L3A
 
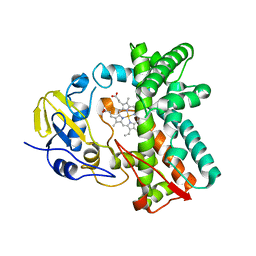 | | Cytochrome P450 107G1 (RapN) with everolimus | | 分子名称: | Cytochrome P450, Everolimus, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Km, V.C, Kim, D.H, Lim, Y.R, Lee, I.H, Lee, J.H, Kang, L.W. | | 登録日 | 2019-10-10 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural insights into CYP107G1 from rapamycin-producing Streptomyces rapamycinicus.
Arch.Biochem.Biophys., 692, 2020
|
|
6L39
 
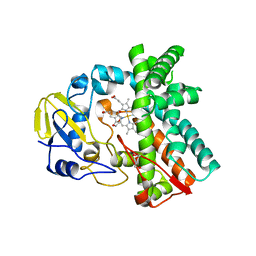 | | Cytochrome P450 107G1 (RapN) | | 分子名称: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | 著者 | Kim, V.C, Kim, D.H, Lim, Y.R, Lee, I.H, Lee, J.H, Kang, L.W. | | 登録日 | 2019-10-10 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.97 Å) | | 主引用文献 | Structural insights into CYP107G1 from rapamycin-producing Streptomyces rapamycinicus.
Arch.Biochem.Biophys., 692, 2020
|
|
7MEM
 
 | |
2V89
 
 | |
6S8N
 
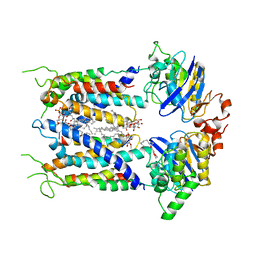 | | Cryo-EM structure of LptB2FGC in complex with lipopolysaccharide | | 分子名称: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, Inner membrane protein yjgQ, ... | | 著者 | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | 登録日 | 2019-07-10 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
6S8H
 
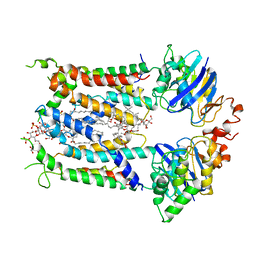 | | Cryo-EM structure of LptB2FG in complex with LPS | | 分子名称: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, ... | | 著者 | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | 登録日 | 2019-07-10 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
8C76
 
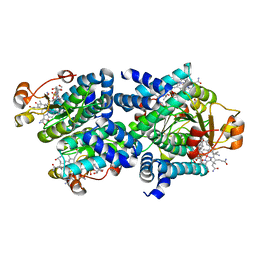 | | Light-state 2.5 Angstrom wild-type X-ray crystal structure of the cobalamin binding domain belonging to a light-dependent transcription regulator TtCarH obtained under aerobic conditions from a form 2 crystal illuminated during 5 s | | 分子名称: | COBALAMIN, Probable transcriptional regulator | | 著者 | Rios-Santacruz, R, Colletier, J.P, Schiro, G, Weik, M. | | 登録日 | 2023-01-12 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Redox driven B 12 -ligand switch drives CarH photoresponse.
Nat Commun, 14, 2023
|
|
8C73
 
 | | Dark state 1.8 Angstrom crystal structure of cobalamin binding domain belonging to a light-dependent transcription regulator TtCarH obtained under aerobic condition form ll | | 分子名称: | 5'-DEOXYADENOSINE, COBALAMIN, Probable transcriptional regulator | | 著者 | Rios-Santacruz, R, Colletier, J.P, Schiro, G, Weik, M. | | 登録日 | 2023-01-12 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Redox driven B 12 -ligand switch drives CarH photoresponse.
Nat Commun, 14, 2023
|
|
8C36
 
 | |
8C34
 
 | |
8C35
 
 | |
8C32
 
 | |
8C33
 
 | |
8C37
 
 | |
8C31
 
 | |
6S8G
 
 | | Cryo-EM structure of LptB2FGC in complex with AMP-PNP | | 分子名称: | Inner membrane protein yjgQ, LPS export ABC transporter permease LptF, Lipopolysaccharide ABC transporter, ... | | 著者 | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | 登録日 | 2019-07-10 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
4QFR
 
 | | Structure of AMPK in complex with Cl-A769662 activator and STAUROSPORINE inhibitor | | 分子名称: | 2-chloro-4-hydroxy-3-(2'-hydroxybiphenyl-4-yl)-6-oxo-6,7-dihydrothieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | 著者 | Calabrese, M.F, Kurumbail, R.G. | | 登録日 | 2014-05-21 | | 公開日 | 2014-08-06 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.34 Å) | | 主引用文献 | Structural Basis for AMPK Activation: Natural and Synthetic Ligands Regulate Kinase Activity from Opposite Poles by Different Molecular Mechanisms.
Structure, 22, 2014
|
|
4QFG
 
 | |
4QFS
 
 | | Structure of AMPK in complex with Br2-A769662core activator and STAUROSPORINE inhibitor | | 分子名称: | 2-bromo-3-(4-bromophenyl)-4-hydroxy-6-oxo-6,7-dihydrothieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | 著者 | Calabrese, M.F, Kurumbail, R.G. | | 登録日 | 2014-05-21 | | 公開日 | 2014-08-06 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.55 Å) | | 主引用文献 | Structural Basis for AMPK Activation: Natural and Synthetic Ligands Regulate Kinase Activity from Opposite Poles by Different Molecular Mechanisms.
Structure, 22, 2014
|
|
6WY1
 
 | | Crystal structure of an engineered thermostable dengue virus 2 envelope protein dimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dengue 2 soluble recombinant envelope | | 著者 | Kudlacek, S.T, Lakshmanane, P, Kuhlman, B. | | 登録日 | 2020-05-12 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.42 Å) | | 主引用文献 | Designed, highly expressing, thermostable dengue virus 2 envelope protein dimers elicit quaternary epitope antibodies.
Sci Adv, 7, 2021
|
|
1TBJ
 
 | | H141A mutant of rat liver arginase I | | 分子名称: | Arginase 1, GLYCEROL, MANGANESE (II) ION | | 著者 | Cama, E, Cox, J.D, Ash, D.E, Christianson, D.W. | | 登録日 | 2004-05-20 | | 公開日 | 2005-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Probing the role of the hyper-reactive histidine residue of arginase.
Arch.Biochem.Biophys., 444, 2005
|
|
