4Q65
 
 | | Structure of the E. coli Peptide Transporter YbgH | | 分子名称: | Dipeptide permease D | | 著者 | Zhang, C, Zhao, Y, Mao, G, Liu, M, Wang, X. | | 登録日 | 2014-04-21 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Crystal structure of the E. coli peptide transporter YbgH.
Structure, 22, 2014
|
|
3RX2
 
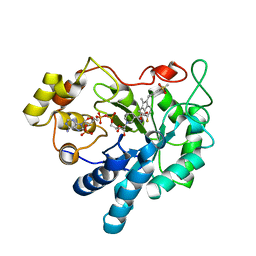 | | Crystal Structure of Human Aldose Reductase Complexed with Sulindac Sulfone | | 分子名称: | 2-[(3Z)-6-fluoranyl-2-methyl-3-[(4-methylsulfonylphenyl)methylidene]inden-1-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zheng, X, Chen, J, Luo, H, Hu, X. | | 登録日 | 2011-05-10 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The molecular basis for inhibition of sulindac and its metabolites towards human aldose reductase
Febs Lett., 586, 2012
|
|
7E24
 
 | |
7E28
 
 | |
7E3X
 
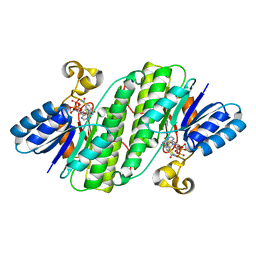 | | Crystal structure of SDR family NAD(P)-dependent oxidoreductase from exiguobacterium | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | 著者 | Chen, L, Tang, J, Yuan, S, Zhang, F, Chen, S. | | 登録日 | 2021-02-09 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structure-guided evolution of a ketoreductase forefficient and stereoselective bioreduction of bulkyalpha-aminobeta-keto esters
Catalysis Science And Technology, 2021
|
|
3RX3
 
 | | Crystal Structure of Human Aldose Reductase Complexed with Sulindac | | 分子名称: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [(1Z)-5-fluoro-2-methyl-1-{4-[methylsulfinyl]benzylidene}-1H-inden-3-yl]acetic acid | | 著者 | Zheng, X, Chen, J, Luo, H, Hu, X. | | 登録日 | 2011-05-10 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The molecular basis for inhibition of sulindac and its metabolites towards human aldose reductase.
Febs Lett., 586, 2012
|
|
4HNE
 
 | | Crystal structure of the catalytic domain of human type II alpha Phosphatidylinositol 4-kinase (PI4KIIalpha) in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha | | 著者 | Zhou, Q, Zhai, Y, Zhang, K, Chen, C, Sun, F. | | 登録日 | 2012-10-19 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Molecular insights into the membrane-associated phosphatidylinositol 4-kinase II alpha.
Nat Commun, 5, 2014
|
|
7ECW
 
 | | The Csy-AcrIF14-dsDNA complex | | 分子名称: | 54-MER DNA, AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, ... | | 著者 | Zhang, L.X, Feng, Y. | | 登録日 | 2021-03-13 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
7ECV
 
 | | The Csy-AcrIF14 complex | | 分子名称: | AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | 著者 | Zhang, L.X, Feng, Y. | | 登録日 | 2021-03-13 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.43 Å) | | 主引用文献 | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
7DU0
 
 | |
7KKZ
 
 | |
7KMD
 
 | |
7KLC
 
 | |
8C0Y
 
 | |
4MXA
 
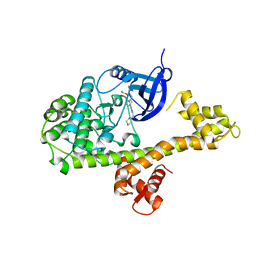 | | CDPK1 from Neospora caninum in complex with inhibitor RM-1-132 | | 分子名称: | 3-(6-ethoxynaphthalen-2-yl)-1-(piperidin-4-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-like domain protein kinase isoenzyme gamma, related | | 著者 | Merritt, E.A. | | 登録日 | 2013-09-26 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Neospora caninum Calcium-Dependent Protein Kinase 1 Is an Effective Drug Target for Neosporosis Therapy.
Plos One, 9, 2014
|
|
6J11
 
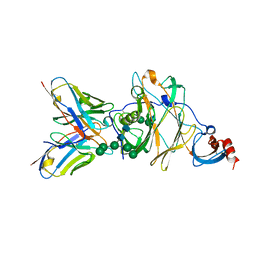 | | MERS-CoV spike N-terminal domain and 7D10 scFv complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-terminal domain of Spike glycoprotein, ... | | 著者 | Zhou, H, Zhang, S, Zhang, S, Tang, W, Wang, X. | | 登録日 | 2018-12-27 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural definition of a neutralization epitope on the N-terminal domain of MERS-CoV spike glycoprotein.
Nat Commun, 10, 2019
|
|
7END
 
 | |
7EN9
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-02 | | 分子名称: | 3C-like proteinase, 5-bromanyl-~{N}-methyl-3-nitro-2-[(4~{R},5~{S})-2-(7-oxidanylisoquinolin-4-yl)carbonyl-4-phenyl-2,7-diazaspiro[4.4]nonan-7-yl]benzamide | | 著者 | Hou, N, Peng, C, Hu, Q. | | 登録日 | 2021-04-16 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7EN8
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-04 | | 分子名称: | 3C-like proteinase, GLYCEROL, ~{N}-[(1~{S},2~{R})-2-[[4-bromanyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]isoquinoline-4-carboxamide | | 著者 | Hou, N, Peng, C, Hu, Q. | | 登録日 | 2021-04-16 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7ENE
 
 | |
5D6S
 
 | | Structure of epoxyqueuosine reductase from Streptococcus thermophilus. | | 分子名称: | COBALAMIN, Epoxyqueuosine reductase, IRON/SULFUR CLUSTER | | 著者 | Payne, K.A.P, Fisher, K, Dunstan, M.S, Sjuts, H, Leys, D. | | 登録日 | 2015-08-12 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Epoxyqueuosine Reductase Structure Suggests a Mechanism for Cobalamin-dependent tRNA Modification.
J.Biol.Chem., 290, 2015
|
|
7NRU
 
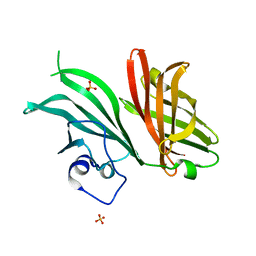 | |
3T8R
 
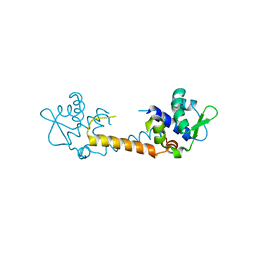 | | Crystal structure of Staphylococcus aureus CymR | | 分子名称: | Staphylococcus aureus CymR | | 著者 | He, C, Ji, Q. | | 登録日 | 2011-08-01 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Staphylococcus aureus CymR Is a New Thiol-based Oxidation-sensing Regulator of Stress Resistance and Oxidative Response.
J.Biol.Chem., 287, 2012
|
|
4MX9
 
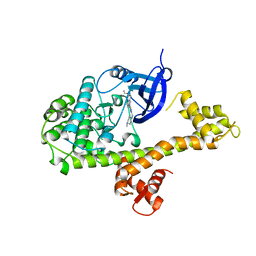 | | CDPK1 from Neospora caninum in complex with inhibitor UW1294 | | 分子名称: | 3-(6-ethoxynaphthalen-2-yl)-1-[(1-methylpiperidin-4-yl)methyl]-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-like domain protein kinase isoenzyme gamma, related | | 著者 | Merritt, E.A. | | 登録日 | 2013-09-26 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Neospora caninum Calcium-Dependent Protein Kinase 1 Is an Effective Drug Target for Neosporosis Therapy.
Plos One, 9, 2014
|
|
7FIV
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip(Tunis) | | 分子名称: | CidA_I gamma/2 protein, CidB_I b/2 protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
