8WKS
 
 | |
8WKX
 
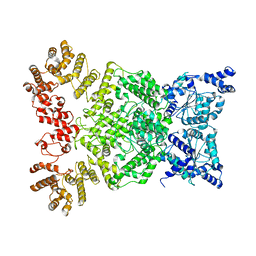 | | Cryo-EM structure of DSR2 | | 分子名称: | SIR2-like domain-containing protein | | 著者 | Gao, A, Huang, J, Zhu, K. | | 登録日 | 2023-09-28 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.15 Å) | | 主引用文献 | Molecular basis of bacterial DSR2 anti-phage defense and viral immune evasion.
Nat Commun, 15, 2024
|
|
8WKT
 
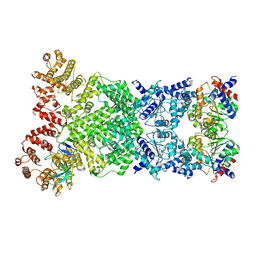 | | Cryo-EM structure of DSR2-DSAD1 complex | | 分子名称: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | 著者 | Gao, A, Huang, J, Zhu, K. | | 登録日 | 2023-09-28 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.86 Å) | | 主引用文献 | Molecular basis of bacterial DSR2 anti-phage defense and viral immune evasion.
Nat Commun, 15, 2024
|
|
2LA8
 
 | |
2L91
 
 | |
8HR7
 
 | | Structure of RdrA-RdrB complex | | 分子名称: | Adenosine deaminase, Archaeal ATPase | | 著者 | Gao, Y. | | 登録日 | 2022-12-15 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.96 Å) | | 主引用文献 | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRC
 
 | | Structure of dodecameric RdrB cage | | 分子名称: | Adenosine deaminase | | 著者 | Gao, Y. | | 登録日 | 2022-12-15 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (2.58 Å) | | 主引用文献 | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRB
 
 | | Structure of tetradecameric RdrA ring in RNA-loading state | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase, RNA (5'-R(*GP*UP*CP*CP*AP*GP*CP*GP*UP*CP*AP*UP*CP*GP*CP*UP*GP*GP*AP*C)-3') | | 著者 | Gao, Y. | | 登録日 | 2022-12-15 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HR8
 
 | | Structure of heptameric RdrA ring | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase | | 著者 | Gao, Y. | | 登録日 | 2022-12-15 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HR9
 
 | | Structure of tetradecameric RdrA ring | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase | | 著者 | Gao, Y. | | 登録日 | 2022-12-15 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRA
 
 | | Structure of heptameric RdrA ring in RNA-loading state | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase, RNA (5'-R(P*GP*UP*CP*CP*AP*GP*CP*GP*UP*CP*AP*UP*CP*GP*CP*UP*GP*GP*AP*C)-3') | | 著者 | Gao, Y. | | 登録日 | 2022-12-15 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HMV
 
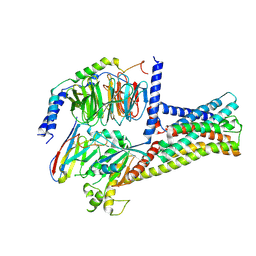 | | Structure of GPR21-Gs complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Wong, T.S, Gao, W. | | 登録日 | 2022-12-05 | | 公開日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Cryo-EM structure of orphan G protein-coupled receptor GPR21.
MedComm (2020), 4, 2023
|
|
8JFG
 
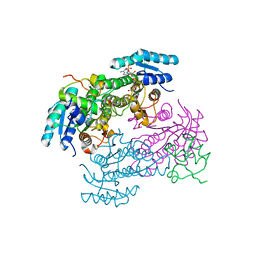 | |
8JFJ
 
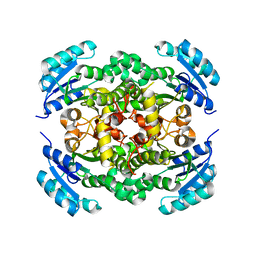 | |
8JFN
 
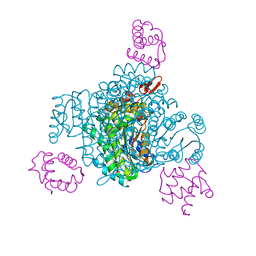 | |
8JFM
 
 | |
8JFI
 
 | |
8JFA
 
 | |
8JFH
 
 | |
8JF9
 
 | |
8IRL
 
 | | Apo state of Arabidopsis AZG1 at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | 著者 | Xu, L, Guo, J. | | 登録日 | 2023-03-19 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRN
 
 | | 6-BAP bound state of Arabidopsis AZG1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1, N-BENZYL-9H-PURIN-6-AMINE | | 著者 | Xu, L, Guo, J. | | 登録日 | 2023-03-19 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRP
 
 | | kinetin bound state of Arabidopsis AZG1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1, N-(FURAN-2-YLMETHYL)-7H-PURIN-6-AMINE | | 著者 | Xu, L, Guo, J. | | 登録日 | 2023-03-19 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRM
 
 | |
8IRO
 
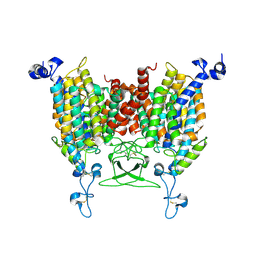 | | trans-Zeatin bound state of Arabidopsis AZG1 at pH7.4 | | 分子名称: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | 著者 | Xu, L, Guo, J. | | 登録日 | 2023-03-19 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
