6MIN
 
 | |
6MIU
 
 | |
6MIM
 
 | |
6MIQ
 
 | | Crystal structure of Taf14 YEATS domain in complex with histone H3K9bu | | 分子名称: | Histone H3K9bu, Transcription initiation factor TFIID subunit 14 | | 著者 | Klein, B.J, Andrews, F.H, Vann, K.R, Kutateladze, T.G. | | 登録日 | 2018-09-19 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
6MIO
 
 | | Crystal structure of Taf14 YEATS domain in complex with histone H3K9pr | | 分子名称: | Histone H3K9pr, Transcription initiation factor TFIID subunit 14 | | 著者 | Klein, B.J, Andrews, F.H, Vann, K.R, Kutateladze, T.G. | | 登録日 | 2018-09-19 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
6MIP
 
 | |
6MJ7
 
 | | Crystal structure of p62 ZZ domain in complex with free arginine | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, ARGININE, Sequestosome-1, ... | | 著者 | Ahn, J, Zhang, Y, Kutateladze, T.G. | | 登録日 | 2018-09-20 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.412 Å) | | 主引用文献 | ZZ-dependent regulation of p62/SQSTM1 in autophagy.
Nat Commun, 9, 2018
|
|
6MIL
 
 | | Crystal structure of AF9 YEATS domain in complex with histone H3K9bu | | 分子名称: | DI(HYDROXYETHYL)ETHER, Histone H3K9bu, MALONATE ION, ... | | 著者 | Vann, K.R, Klein, B.J, Kutateladze, T.G. | | 登録日 | 2018-09-19 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
6O7G
 
 | |
1P39
 
 | |
6JEP
 
 | | Structure of a neutralizing antibody bound to the Zika envelope protein domain III | | 分子名称: | Genome polyprotein, heavy chain of Fab ZK2B10, light chain of Fab ZK2B10 | | 著者 | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | 登録日 | 2019-02-07 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.316 Å) | | 主引用文献 | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
6JFI
 
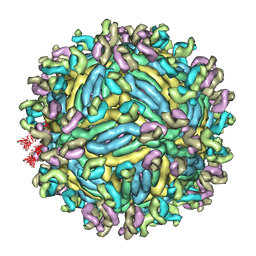 | | The symmetric-reconstructed cryo-EM structure of Zika virus-FabZK2B10 complex | | 分子名称: | FabZK2B10 heavy chain, FabZK2B10 light chain, ZIKV structural protein E, ... | | 著者 | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | 登録日 | 2019-02-08 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (11 Å) | | 主引用文献 | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
6JFH
 
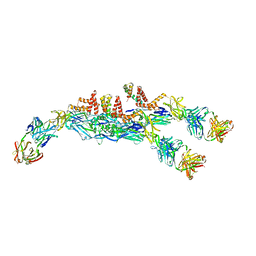 | | The asymmetric-reconstructed cryo-EM structure of Zika virus-FabZK2B10 complex | | 分子名称: | FabZK2B10 heavy chain, FabZK2B10 light chain, ZIKV structural E protein, ... | | 著者 | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | 登録日 | 2019-02-08 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (20 Å) | | 主引用文献 | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
5UQ9
 
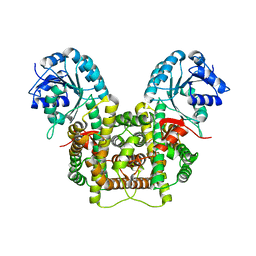 | | Crystal structure of 6-phosphogluconate dehydrogenase with ((4R,5R)-5-(hydroxycarbamoyl)-2,2-dimethyl-1,3-dioxolan-4-yl)methyl dihydrogen phosphate | | 分子名称: | 6-phosphogluconate dehydrogenase, decarboxylating, [(4R,5R)-5-(hydroxycarbamoyl)-2,2-dimethyl-1,3-dioxolan-4-yl]methyl dihydrogen phosphate | | 著者 | Leonard, P.G. | | 登録日 | 2017-02-07 | | 公開日 | 2018-08-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Functional Genomics Reveals Synthetic Lethality between Phosphogluconate Dehydrogenase and Oxidative Phosphorylation.
Cell Rep, 26, 2019
|
|
7XS2
 
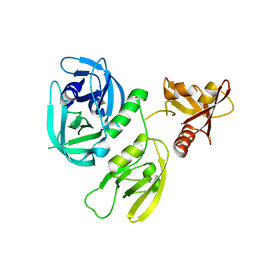 | | Monomer structure of HtrA from Helicobacter pylori | | 分子名称: | Periplasmic serine endoprotease DegP-like | | 著者 | Cui, L, Liu, W. | | 登録日 | 2022-05-12 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structures and solution conformations of HtrA from Helicobacter pylori reveal pH-dependent oligomeric conversion and conformational rearrangements.
Int.J.Biol.Macromol., 243, 2023
|
|
7XS0
 
 | |
5ZXV
 
 | |
6DCE
 
 | | X-ray structure of FIP200 claw domain | | 分子名称: | RB1-inducible coiled-coil protein 1, SULFATE ION | | 著者 | Su, M.-Y, Hurley, J.H. | | 登録日 | 2018-05-05 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates.
Mol. Cell, 74, 2019
|
|
5YY5
 
 | | Structural definition of a unique neutralization epitope on the receptor-binding domain of MERS-CoV spike glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, Light chain, ... | | 著者 | Zhang, S, Wang, P, Zhou, P, Wang, X, Zhang, L. | | 登録日 | 2017-12-08 | | 公開日 | 2018-08-01 | | 最終更新日 | 2025-04-09 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Definition of a Unique Neutralization Epitope on the Receptor-Binding Domain of MERS-CoV Spike Glycoprotein
Cell Rep, 24, 2018
|
|
6DVK
 
 | | Computationally designed mini tetraloop-tetraloop receptor by the RNAMake program - construct 6 (miniTTR 6) | | 分子名称: | COBALT (II) ION, MAGNESIUM ION, RNA (95-MER) | | 著者 | Eiler, D.R, Yesselman, J.D, Costantino, D.A, Das, R, Kieft, J.S. | | 登録日 | 2018-06-24 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Computational design of three-dimensional RNA structure and function.
Nat Nanotechnol, 14, 2019
|
|
3O35
 
 | |
5HJB
 
 | | AF9 YEATS in complex with histone H3 Crotonylation at K9 | | 分子名称: | Protein AF-9, peptide of Histone H3.1 | | 著者 | Li, Y.Y, Zhao, D, Guan, H.P, Li, H.T. | | 登録日 | 2016-01-12 | | 公開日 | 2016-04-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain
Mol.Cell, 62, 2016
|
|
5HJD
 
 | | AF9 YEATS in complex with histone H3 Crotonylation at K18 | | 分子名称: | COPPER (II) ION, Protein AF-9, SULFATE ION, ... | | 著者 | Li, Y.Y, Zhao, D, Guan, H.P, Li, H.T. | | 登録日 | 2016-01-13 | | 公開日 | 2016-04-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.806 Å) | | 主引用文献 | Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain.
Mol.Cell, 62, 2016
|
|
5HJC
 
 | | BRD3 second bromodomain in complex with histone H3 acetylation at K18 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, CHLORIDE ION, ... | | 著者 | Li, Y.Y, Zhao, D, Guan, H.P, Li, H.T. | | 登録日 | 2016-01-13 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain
Mol.Cell, 62, 2016
|
|
4N4I
 
 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.3K36me3 | | 分子名称: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.3, ... | | 著者 | Li, Y, Ren, Y, Li, H. | | 登録日 | 2013-10-08 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
