7D26
 
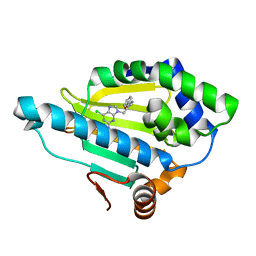 | | Hsp90 alpha N-terminal domain in complex with a 8 compund | | 分子名称: | 6-chloranyl-9-[(2-phenyl-1,3-oxazol-5-yl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | 著者 | Shin, S.C, Kim, E.E. | | 登録日 | 2020-09-15 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D22
 
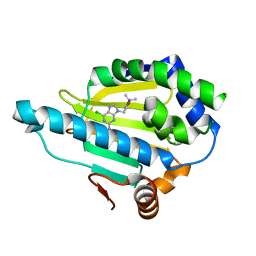 | | Hsp90 alpha N-terminal domain in complex with a 6B compund | | 分子名称: | 9-[(3-tert-butyl-1,2-oxazol-5-yl)methyl]-6-chloranyl-purin-2-amine, Heat shock protein HSP 90-alpha | | 著者 | Shin, S.C, Kim, E.E. | | 登録日 | 2020-09-15 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D25
 
 | |
7D1V
 
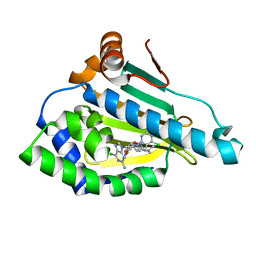 | | Hsp90 alpha N-terminal domain in complex with a 6C compund | | 分子名称: | 6-chloranyl-9-[(3-propan-2-yl-1,2-oxazol-5-yl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | 著者 | Shin, S.C, Kim, E.E. | | 登録日 | 2020-09-15 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.332 Å) | | 主引用文献 | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
8JQZ
 
 | | Crystal Structure of GppNHp-bound mIRGB10 | | 分子名称: | Immunity-related GTPase family member b10, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Ha, H.J, Park, H.H. | | 登録日 | 2023-06-15 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structural basis of IRGB10 oligomerization by GTP hydrolysis.
Front Immunol, 14, 2023
|
|
8JQY
 
 | |
6M36
 
 | |
7EEW
 
 | |
3HB1
 
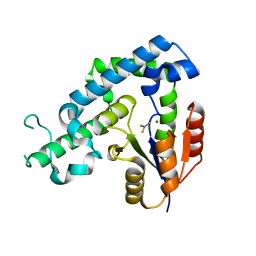 | | Crystal structure of ed-eya2 complexed with Alf3 | | 分子名称: | ALUMINUM FLUORIDE, Eyes absent homolog 2 (Drosophila), MAGNESIUM ION | | 著者 | Jung, S.K, Jeong, D.G, Ryu, S.E, Kim, S.J. | | 登録日 | 2009-05-03 | | 公開日 | 2009-12-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Crystal structure of ED-Eya2: insight into dual roles as a protein tyrosine phosphatase and a transcription factor
Faseb J., 24, 2010
|
|
3HB0
 
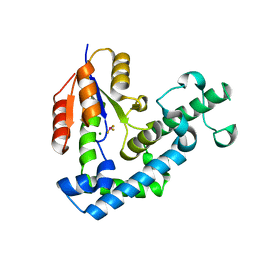 | | Structure of edeya2 complexed with bef3 | | 分子名称: | BERYLLIUM TRIFLUORIDE ION, Eyes absent homolog 2 (Drosophila), MAGNESIUM ION | | 著者 | Jung, S.K, Jeong, D.G, Ryu, S.E, Kim, S.J. | | 登録日 | 2009-05-03 | | 公開日 | 2009-12-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of ED-Eya2: insight into dual roles as a protein tyrosine phosphatase and a transcription factor
Faseb J., 24, 2010
|
|
8U4N
 
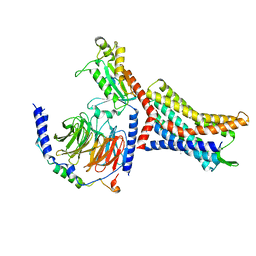 | |
8U4S
 
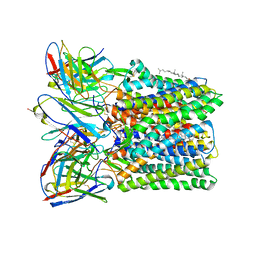 | |
8U4T
 
 | |
8U4P
 
 | |
8U4Q
 
 | |
8U4R
 
 | |
8U4O
 
 | |
3CMJ
 
 | |
6TY0
 
 | |
6TY2
 
 | |
7BOL
 
 | |
4R3Z
 
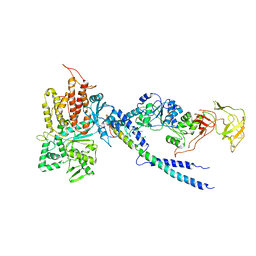 | | Crystal structure of human ArgRS-GlnRS-AIMP1 complex | | 分子名称: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 1, Arginine--tRNA ligase, cytoplasmic, ... | | 著者 | Fu, Y, Kim, Y, Cho, Y. | | 登録日 | 2014-08-18 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (4.033 Å) | | 主引用文献 | Structure of the ArgRS-GlnRS-AIMP1 complex and its implications for mammalian translation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5HZU
 
 | | Crystal structure of Dronpa-Ni2+ | | 分子名称: | Fluorescent protein Dronpa, NICKEL (II) ION | | 著者 | Hwang, K.Y, Nam, K.H. | | 登録日 | 2016-02-03 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
FEBS Lett., 590, 2016
|
|
5HZS
 
 | | Crystal structure of Dronpa-Co2+ | | 分子名称: | COBALT (II) ION, Fluorescent protein Dronpa | | 著者 | Hwang, K.Y, Nam, K.H. | | 登録日 | 2016-02-03 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
FEBS Lett., 590, 2016
|
|
5HZT
 
 | | Crystal structure of Dronpa-Cu2+ | | 分子名称: | COPPER (II) ION, Fluorescent protein Dronpa | | 著者 | Hwang, K.Y, Nam, K.H. | | 登録日 | 2016-02-03 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
FEBS Lett., 590, 2016
|
|
