5N70
 
 | | CRYSTAL STRUCTURE OF MATURE CATHEPSIN D FROM THE TICK IXODES RICINUS (IRCD1) IN COMPLEX WITH THE N-TERMINAL OCTAPEPTIDE OF THE PROPEPTID | | 分子名称: | ALA-PHE-ARG-ILE-PRO-LEU-THR-ARG, Putative cathepsin d | | 著者 | Brynda, J, Hanova, I, Hobizalova, R, Mares, M. | | 登録日 | 2017-02-17 | | 公開日 | 2017-12-27 | | 最終更新日 | 2018-03-28 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Novel Structural Mechanism of Allosteric Regulation of Aspartic Peptidases via an Evolutionarily Conserved Exosite.
Cell Chem Biol, 25, 2018
|
|
5N7N
 
 | | CRYSTAL STRUCTURE OF CATHEPSIN D ZYMOGEN FROM THE TICK IXODES RICINUS (IRCD1) | | 分子名称: | AMMONIUM ION, Putative cathepsin d, SULFATE ION | | 著者 | Brynda, J, Hanova, I, Hobizalova, R, Mares, M. | | 登録日 | 2017-02-21 | | 公開日 | 2017-12-27 | | 最終更新日 | 2018-03-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Novel Structural Mechanism of Allosteric Regulation of Aspartic Peptidases via an Evolutionarily Conserved Exosite.
Cell Chem Biol, 25, 2018
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | 登録日 | 2022-07-05 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (4.17 Å) | | 主引用文献 | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7FBJ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing nanobody 17F6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, ... | | 著者 | Zhu, J, Xu, T, Feng, B, Liu, J. | | 登録日 | 2021-07-11 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
7FBK
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain N501Y mutant in complex with neutralizing nanobody 20G6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, Spike protein S1 | | 著者 | Zhu, J, Xu, T, Feng, B, Liu, J. | | 登録日 | 2021-07-11 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
7XM5
 
 | | Keap1 Kelch domain (residues 322-609) in complex with 6i | | 分子名称: | Kelch-like ECH-associated protein 1, N-[4-[(2-azanyl-2-oxidanylidene-ethyl)-[4-[(2-azanyl-2-oxidanylidene-ethyl)-(4-methoxyphenyl)sulfonyl-amino]naphthalen-1-yl]sulfamoyl]phenyl]-3-morpholin-4-yl-propanamide | | 著者 | Xu, K. | | 登録日 | 2022-04-24 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystallography-Guided Optimizations of the Keap1-Nrf2 Inhibitors on the Solvent Exposed Region: From Symmetric to Asymmetric Naphthalenesulfonamides.
J.Med.Chem., 65, 2022
|
|
7XM4
 
 | | Crystal structure of Keap1 Kelch domain (residues 322-609) in complex with 6e | | 分子名称: | Kelch-like ECH-associated protein 1, N-[4-[(2-azanyl-2-oxidanylidene-ethyl)-[4-[(2-azanyl-2-oxidanylidene-ethyl)-(4-methoxyphenyl)sulfonyl-amino]naphthalen-1-yl]sulfamoyl]phenyl]-2-(4-ethylpiperazin-1-yl)ethanamide | | 著者 | Xu, K. | | 登録日 | 2022-04-24 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystallography-Guided Optimizations of the Keap1-Nrf2 Inhibitors on the Solvent Exposed Region: From Symmetric to Asymmetric Naphthalenesulfonamides.
J.Med.Chem., 65, 2022
|
|
7XM3
 
 | | Crystal structure of Keap1 Kelch domain (residues 322-609) in complex with 6k | | 分子名称: | Kelch-like ECH-associated protein 1, N-[4-[(2-azanyl-2-oxidanylidene-ethyl)-[4-[(2-azanyl-2-oxidanylidene-ethyl)-(4-methoxyphenyl)sulfonyl-amino]naphthalen-1-yl]sulfamoyl]phenyl]-3-(4-ethylpiperazin-1-yl)propanamide | | 著者 | Xu, K. | | 登録日 | 2022-04-24 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystallography-Guided Optimizations of the Keap1-Nrf2 Inhibitors on the Solvent Exposed Region: From Symmetric to Asymmetric Naphthalenesulfonamides.
J.Med.Chem., 65, 2022
|
|
7XM2
 
 | | Crystal structure of Keap1 Kelch domain (residues 322-609) in complex with NXPZ-2 | | 分子名称: | 2-[(4-aminophenyl)sulfonyl-[4-[(2-azanyl-2-oxidanylidene-ethyl)-(4-methoxyphenyl)sulfonyl-amino]naphthalen-1-yl]amino]ethanamide, Kelch-like ECH-associated protein 1 | | 著者 | Xu, K. | | 登録日 | 2022-04-24 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystallography-Guided Optimizations of the Keap1-Nrf2 Inhibitors on the Solvent Exposed Region: From Symmetric to Asymmetric Naphthalenesulfonamides.
J.Med.Chem., 65, 2022
|
|
7R7Z
 
 | | Crystal structure of QW9-HLA-B*5301 specific T Cell Receptor, C3 | | 分子名称: | Alpha chain of C3 TCR, Beta chain of C3 TCR, HEXAETHYLENE GLYCOL, ... | | 著者 | Li, X.L, Tan, K.M, Xu, S.T, Ng, R, Walker, B.D, Wang, J.H. | | 登録日 | 2021-06-25 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.286 Å) | | 主引用文献 | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R7Y
 
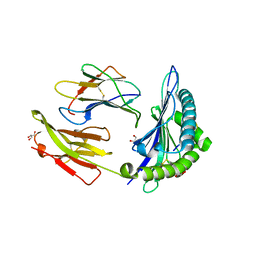 | | Crystal structure of HLA-B*5701 complex with an HIV-1 Gag-derived epitope QW9 S3T variant | | 分子名称: | Beta-2-microglobulin, GLN-ALA-THR-GLN-GLU-VAL-LYS-ASN-TRP, GLYCEROL, ... | | 著者 | Li, X.L, Ng, R, Tan, K.M, Walker, B.D, Wang, J.H. | | 登録日 | 2021-06-25 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.601 Å) | | 主引用文献 | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R7V
 
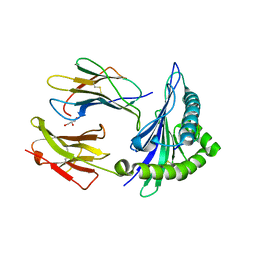 | | Crystal structure of HLA-B*5301 complex with an HIV-1 Gag-derived epitope QW9 | | 分子名称: | Beta-2-microglobulin, GLN-ALA-SER-GLN-GLU-VAL-LYS-ASN-TRP, GLYCEROL, ... | | 著者 | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | 登録日 | 2021-06-25 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R7W
 
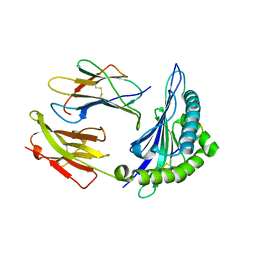 | | Crystal structure of HLA-B*5301 complex with an HIV-1 Gag-derived epitope QW9 S3T variant | | 分子名称: | Beta-2-microglobulin, GLN-ALA-THR-GLN-GLU-VAL-LYS-ASN-TRP, MHC class I antigen | | 著者 | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | 登録日 | 2021-06-25 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R7X
 
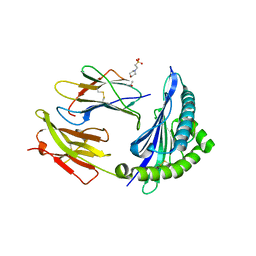 | | Crystal structure of HLA-B*5701 complex with an HIV-1 Gag-derived epitope QW9 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, GLN-ALA-SER-GLN-GLU-VAL-LYS-ASN-TRP, ... | | 著者 | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | 登録日 | 2021-06-25 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R80
 
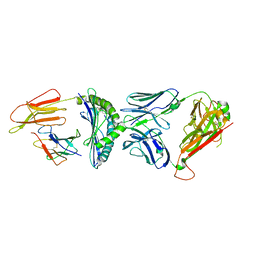 | | Crystal structure of C3 TCR complex with QW9-bound HLA-B*5301 | | 分子名称: | Alpha chain of C3 TCR, Beta Chain of C3 TCR, Beta-2-microglobulin, ... | | 著者 | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | 登録日 | 2021-06-25 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
6KXG
 
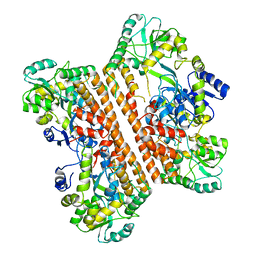 | |
6KI0
 
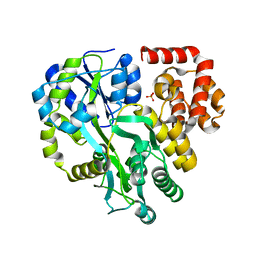 | | Crystal Structure of Human ASC-CARD | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Xu, Z.H, Jin, T.C. | | 登録日 | 2019-07-16 | | 公開日 | 2020-07-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Homotypic CARD-CARD interaction is critical for the activation of NLRP1 inflammasome.
Cell Death Dis, 12, 2021
|
|
5KCC
 
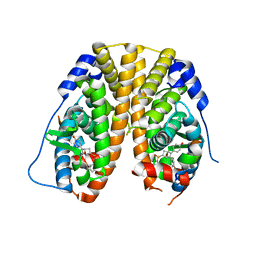 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with Oxabicyclic Heptene Sulfonamide (OBHS-N) | | 分子名称: | (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-N-phenyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2016-06-06 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.386 Å) | | 主引用文献 | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5GPP
 
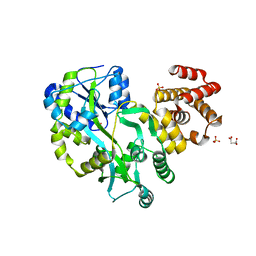 | | Crystal structure of zebrafish ASC PYD domain | | 分子名称: | ACETATE ION, Maltose-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, ... | | 著者 | Jin, T, Li, Y. | | 登録日 | 2016-08-04 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Functional and structural characterization of zebrafish ASC.
FEBS J., 285, 2018
|
|
5GPQ
 
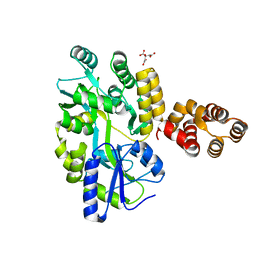 | | Crystal Structure of zebrafish ASC CARD Domain | | 分子名称: | CITRIC ACID, Maltose-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Jin, T, Li, Y. | | 登録日 | 2016-08-04 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Functional and structural characterization of zebrafish ASC.
FEBS J., 285, 2018
|
|
5HG9
 
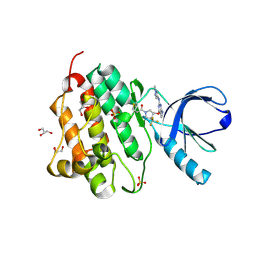 | | EGFR (L858R, T790M, V948R) in complex with 1-[(3R,4R)-3-[({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-(trifluoromethyl)pyrrolidin-1-yl]prop-2-en-1-one | | 分子名称: | 1-[(3R,4R)-3-[({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-(trifluoromethyl)pyrrolidin-1-yl]propan-1-one, Epidermal growth factor receptor, GLYCEROL, ... | | 著者 | Gajiwala, K.S. | | 登録日 | 2016-01-08 | | 公開日 | 2016-02-03 | | 最終更新日 | 2016-03-23 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
5HG8
 
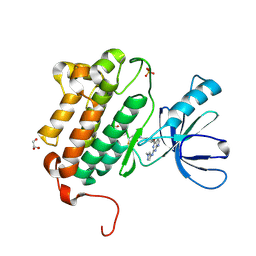 | | EGFR (L858R, T790M, V948R) in complex with N-[3-({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)phenyl]prop-2-enamide | | 分子名称: | Epidermal growth factor receptor, GLYCEROL, N-[3-({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)phenyl]propanamide, ... | | 著者 | Gajiwala, K.S. | | 登録日 | 2016-01-08 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
7C8U
 
 | | The crystal structure of COVID-19 main protease in complex with GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Luan, X, Shang, W, Wang, Y, Yin, W, Jiang, Y, Feng, S, Wang, Y, Liu, M, Zhou, R, Zhang, Z, Wang, F, Cheng, W, Gao, M, Wang, H, Wu, W, Tian, R, Tian, Z, Jin, Y, Jiang, H.W, Zhang, L, Xu, H.E, Zhang, S. | | 登録日 | 2020-06-03 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The crystal structure of COVID-19 main protease in complex with GC376
To Be Published
|
|
8VHE
 
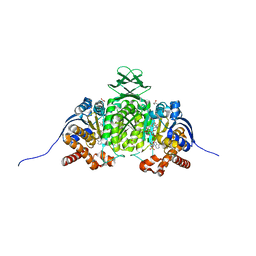 | | Crystal Structure of Human IDH1 R132Q in Complex with NADPH-TCEP Adduct | | 分子名称: | 3,3',3''-({(4R)-1-[(2R,3R,4S,5R)-5-({[(S)-{[(S)-{[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-3,4-dihydroxyoxolan-2-yl]-3-carbamoyl-1,4-dihydropyridin-4-yl}-lambda~5~-phosphanetriyl)tripropanoic acid, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Mealka, M, Sohl, C.D, Huxford, T. | | 登録日 | 2023-12-31 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
8VHB
 
 | | Crystal Structure of Human IDH1 R132Q in complex with NADPH and Alpha-Ketoglutarate | | 分子名称: | (3~{S})-3-[(4~{S})-3-aminocarbonyl-1-[(2~{R},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]-4~{H}-pyridin-4-yl]-2-oxidanylidene-pentanedioic acid, 2-OXOGLUTARIC ACID, CALCIUM ION, ... | | 著者 | Mealka, M, Sohl, C.D, Huxford, T. | | 登録日 | 2023-12-31 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Active site remodeling in tumor-relevant IDH1 mutants drives distinct kinetic features and potential resistance mechanisms.
Nat Commun, 15, 2024
|
|
