8G9U
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | 分子名称: | CRISPR-associated protein, Csd1 family, Csd2 family, ... | | 著者 | Hu, C, Nam, K.H, Ke, A. | | 登録日 | 2023-02-22 | | 公開日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8GAF
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | 分子名称: | Cas11, Cas5, Cas7, ... | | 著者 | Hu, C, Nam, K.H, Ke, A. | | 登録日 | 2023-02-22 | | 公開日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8GAM
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | 分子名称: | Cas11, Cas5, Cas7, ... | | 著者 | Hu, C, Nam, K.H, Ke, A. | | 登録日 | 2023-02-23 | | 公開日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8GAN
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | 分子名称: | Cas11, Cas5, Cas7, ... | | 著者 | Hu, C, Nam, K.H, Ke, A. | | 登録日 | 2023-02-23 | | 公開日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
5HZS
 
 | | Crystal structure of Dronpa-Co2+ | | 分子名称: | COBALT (II) ION, Fluorescent protein Dronpa | | 著者 | Hwang, K.Y, Nam, K.H. | | 登録日 | 2016-02-03 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development
FEBS Lett., 590, 2016
|
|
5HXY
 
 | | Crystal structure of XerA recombinase | | 分子名称: | PHOSPHATE ION, Tyrosine recombinase XerA | | 著者 | Hwang, K.Y, Nam, K.H. | | 登録日 | 2016-01-31 | | 公開日 | 2017-02-01 | | 最終更新日 | 2020-02-19 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of Thermoplasma acidophilum XerA recombinase shows large C-shape clamp conformation and cis-cleavage mode for nucleophilic tyrosine
FEBS Lett., 590, 2016
|
|
6K1G
 
 | | Crystal structure of the L-fucose isomerase soaked with Mn2+ from Raoultella sp. | | 分子名称: | L-fucose isomerase, MANGANESE (II) ION | | 著者 | Kim, I.J, Kim, D.H, Nam, K.H, Kim, K.H. | | 登録日 | 2019-05-10 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.96 Å) | | 主引用文献 | Enzymatic synthesis of l-fucose from l-fuculose using a fucose isomerase fromRaoultellasp. and the biochemical and structural analyses of the enzyme.
Biotechnol Biofuels, 12, 2019
|
|
6K1F
 
 | | Crystal structure of the L-fucose isomerase from Raoultella sp. | | 分子名称: | L-fucose isomerase, MANGANESE (II) ION | | 著者 | Kim, I.J, Kim, D.H, Nam, K.H, Kim, K.H. | | 登録日 | 2019-05-10 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Enzymatic synthesis of l-fucose from l-fuculose using a fucose isomerase fromRaoultellasp. and the biochemical and structural analyses of the enzyme.
Biotechnol Biofuels, 12, 2019
|
|
8D8N
 
 | | gRAMP non-match PFS target RNA | | 分子名称: | RAMP superfamily protein, RNA (35-MER), RNA (5'-R(P*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*AP*CP*A)-3'), ... | | 著者 | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | 登録日 | 2022-06-08 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D9H
 
 | | gRAMP-TPR-CHAT match PFS target RNA(Craspase) | | 分子名称: | CHAT domain protein, PHOSPHATE ION, RAMP superfamily protein, ... | | 著者 | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | 登録日 | 2022-06-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D9E
 
 | | gRAMP-match PFS target | | 分子名称: | RAMP superfamily protein, RNA (36-MER), RNA (5'-R(P*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*GP*UP*A)-3'), ... | | 著者 | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | 登録日 | 2022-06-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D9F
 
 | | gRAMP-TPR-CHAT (Craspase) | | 分子名称: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | 著者 | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | 登録日 | 2022-06-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D97
 
 | | Apo gRAMP | | 分子名称: | RAMP superfamily protein, RNA (42-MER), ZINC ION | | 著者 | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | 登録日 | 2022-06-09 | | 公開日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D9G
 
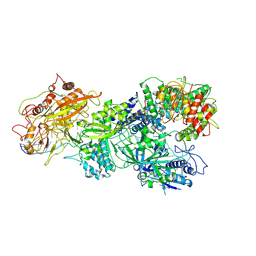 | | gRAMP-TPR-CHAT Non match PFS target RNA(Craspase) | | 分子名称: | CHAT domain protein, RAMP superfamily protein, RNA (36-MER), ... | | 著者 | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | 登録日 | 2022-06-09 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
8D9I
 
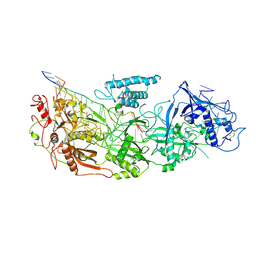 | | gRAMP non-matching PFS-with Mg | | 分子名称: | RAMP superfamily protein, RNA (35-MER), RNA (5'-R(P*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*GP*A)-3'), ... | | 著者 | Hu, C, Nam, K.H, Schuler, G, Ke, A. | | 登録日 | 2022-06-09 | | 公開日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Craspase is a CRISPR RNA-guided, RNA-activated protease.
Science, 377, 2022
|
|
5Z6V
 
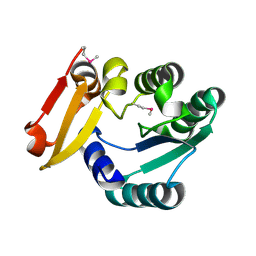 | |
6AA7
 
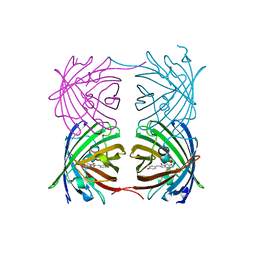 | |
3V9A
 
 | |
3DRE
 
 | |
3DRB
 
 | | Crystal structure of Human Brain-type Creatine Kinase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase B-type, MAGNESIUM ION | | 著者 | Moon, J.H, Bong, S.M, Hwang, K.Y, Chi, Y.M. | | 登録日 | 2008-07-11 | | 公開日 | 2009-03-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural studies of human brain-type creatine kinase complexed with the ADP-Mg2+-NO3- -creatine transition-state analogue complex
Febs Lett., 582, 2008
|
|
5Y9Q
 
 | |
6J43
 
 | | Proteinase K determined by PAL-XFEL | | 分子名称: | CALCIUM ION, Proteinase K | | 著者 | Lee, S.J, Park, J. | | 登録日 | 2019-01-07 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Application of a high-throughput microcrystal delivery system to serial femtosecond crystallography.
J.Appl.Crystallogr., 53, 2020
|
|
7FFT
 
 | |
7FFW
 
 | | The crystal structure of a domain-swapped dimeric maltodextrin-binding protein MalE from Salmonella enterica | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, METHOXYETHANE, ... | | 著者 | Wang, L, Bu, T, Bai, X. | | 登録日 | 2021-07-23 | | 公開日 | 2021-09-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of the domain-swapped dimeric maltodextrin-binding protein MalE from Salmonella enterica.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7MID
 
 | | Sub-complex of Cas4-Cas1-Cas2 bound PAM containing DNA | | 分子名称: | CRISPR-associated endoribonuclease Cas2, CRISPR-associated exonuclease Cas4/endonuclease Cas1 fusion, DNA (33-MER), ... | | 著者 | Hu, C.Y, Ke, A.K. | | 登録日 | 2021-04-16 | | 公開日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.56 Å) | | 主引用文献 | Mechanism for Cas4-assisted directional spacer acquisition in CRISPR-Cas.
Nature, 598, 2021
|
|
