6JHP
 
 | |
5WDK
 
 | | A processive dipeptidyl aminopeptidase secreted from an established commensal bacterium P. distasonis | | 分子名称: | 5-[(3aS,4R,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-N-(2-oxopropyl)pentanamide, Aminopeptidase C, POTASSIUM ION | | 著者 | Wolan, D.W, Xu, J.H, Solania, A, Chatterjee, S, Jiang, Z, ODonoghue, A.J. | | 登録日 | 2017-07-05 | | 公開日 | 2018-07-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | A Commensal Dipeptidyl Aminopeptidase with Specificity for N-Terminal Glycine Degrades Human-Produced Antimicrobial Peptides in Vitro.
Acs Chem.Biol., 13, 2018
|
|
5WDL
 
 | | A processive dipeptidyl aminopeptidase secreted from an established commensal bacterium P. distasonis | | 分子名称: | Aminopeptidase C, N-[(3S)-6-carbamimidamido-2-oxohexan-3-yl]glycinamide, POTASSIUM ION | | 著者 | Wolan, D.W, Xu, J.H, Solania, A, Chatterjee, S, Jiang, Z, ODonoghue, A.J. | | 登録日 | 2017-07-05 | | 公開日 | 2018-07-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.625 Å) | | 主引用文献 | A Commensal Dipeptidyl Aminopeptidase with Specificity for N-Terminal Glycine Degrades Human-Produced Antimicrobial Peptides in Vitro.
Acs Chem.Biol., 13, 2018
|
|
5XGI
 
 | | Crystal structure of PI3K complex with an inhibitor | | 分子名称: | 3-azanyl-5-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | 著者 | Song, K, Yang, X, Zhao, Y, Jian, Z. | | 登録日 | 2017-04-13 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Crystal structure of PI3K complex with an inhibitor
To Be Published
|
|
5XGJ
 
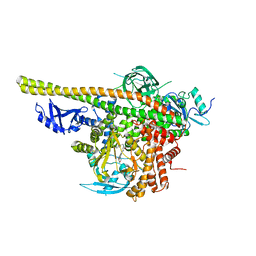 | | Crystal structure of PI3K complex with an inhibitor | | 分子名称: | 3-(4-morpholin-4-ylfuro[3,2-d]pyrimidin-2-yl)-5-[(phenylmethyl)amino]phenol, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | 著者 | Song, K, Yang, X, Zhao, Y, Jian, Z. | | 登録日 | 2017-04-13 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.97 Å) | | 主引用文献 | Crystal structure of PI3K complex with an inhibitor
To Be Published
|
|
7C7Q
 
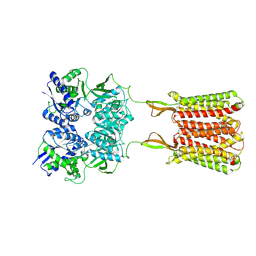 | | Cryo-EM structure of the baclofen/BHFF-bound human GABA(B) receptor in active state | | 分子名称: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | 著者 | Mao, C, Shen, C, Li, C, Shen, D, Xu, C, Zhang, S, Zhou, R, Shen, Q, Chen, L, Jiang, Z, Liu, J, Zhang, Y. | | 登録日 | 2020-05-26 | | 公開日 | 2020-07-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM structures of inactive and active GABABreceptor.
Cell Res., 30, 2020
|
|
7C7S
 
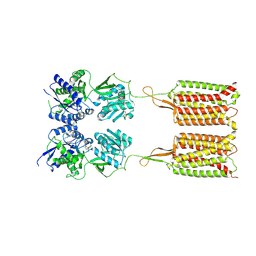 | | Cryo-EM structure of the CGP54626-bound human GABA(B) receptor in inactive state. | | 分子名称: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | 著者 | Mao, C, Shen, C, Li, C, Shen, D, Xu, C, Zhang, S, Zhou, R, Shen, Q, Chen, L, Jiang, Z, Liu, J, Zhang, Y. | | 登録日 | 2020-05-26 | | 公開日 | 2020-07-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM structures of inactive and active GABABreceptor.
Cell Res., 30, 2020
|
|
3NLA
 
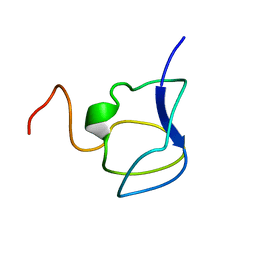 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, 40 STRUCTURES | | 分子名称: | ANTIFREEZE PROTEIN RD3 TYPE III | | 著者 | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | 登録日 | 1998-02-24 | | 公開日 | 1999-02-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
3RDN
 
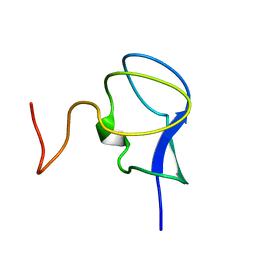 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | ANTIFREEZE PROTEIN RD3 TYPE III | | 著者 | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | 登録日 | 1998-02-24 | | 公開日 | 1999-02-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
7F2E
 
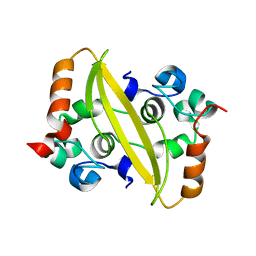 | |
7F2B
 
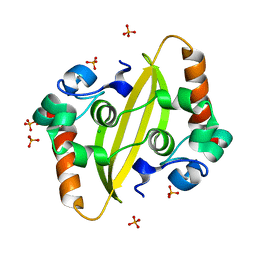 | |
8HXS
 
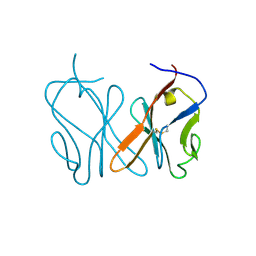 | | Small_spotted catshark CD8alpha | | 分子名称: | T-cell surface glycoprotein CD8 alpha chain | | 著者 | Wang, J, Zou, J. | | 登録日 | 2023-01-05 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | The first crystal structure of CD8 alpha alpha from a cartilaginous fish.
Front Immunol, 14, 2023
|
|
5YUQ
 
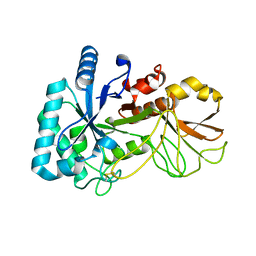 | | The high resolution structure of chitinase (RmChi1) from the thermophilic fungus Rhizomucor miehei (sp P1) | | 分子名称: | Chintase | | 著者 | Jiang, Z.Q, Hu, S.Q, Liu, Y.C, Qin, Z, Yan, Q.J, Yang, S.Q. | | 登録日 | 2017-11-23 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 2021
|
|
6INX
 
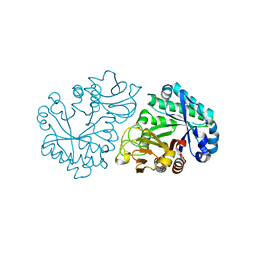 | |
7FBT
 
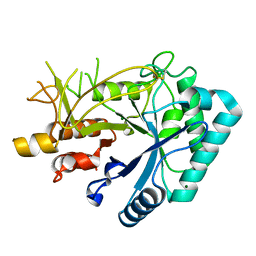 | | Crystal structure of chitinase (RmChi1) from Rhizomucor miehei (sp p32 2 1, MR) | | 分子名称: | Chitinase, MAGNESIUM ION | | 著者 | Jiang, Z.Q, Hu, S.Q, Zhu, Q, Liu, Y.C, Ma, J.W, Yan, Q.J, Gao, Y.G, Yang, S.Q. | | 登録日 | 2021-07-12 | | 公開日 | 2021-08-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
6KJA
 
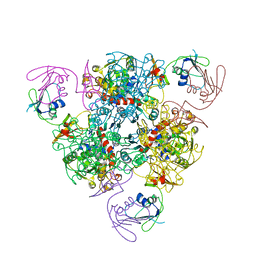 | |
6KJB
 
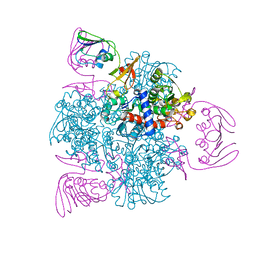 | | wild-type apo-form E. coli ATCase holoenzyme with an unusual open conformation of R167 | | 分子名称: | Aspartate carbamoyltransferase catalytic subunit, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | 著者 | Wang, N, Lei, Z, Zheng, J, Jia, Z.C. | | 登録日 | 2019-07-22 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Conformational Plasticity of the Active Site Entrance inE. coliAspartate Transcarbamoylase and Its Implication in Feedback Regulation.
Int J Mol Sci, 21, 2020
|
|
5MH1
 
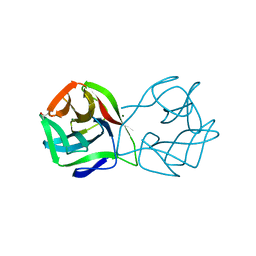 | | Crystal structure of a DM9 domain containing protein from Crassostrea gigas | | 分子名称: | GLYCEROL, MAGNESIUM ION, Natterin-3, ... | | 著者 | Weinert, T, Warkentin, E, Pang, G. | | 登録日 | 2016-11-22 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | DM9 Domain Containing Protein Functions As a Pattern Recognition Receptor with Broad Microbial Recognition Spectrum.
Front Immunol, 8, 2017
|
|
5MH3
 
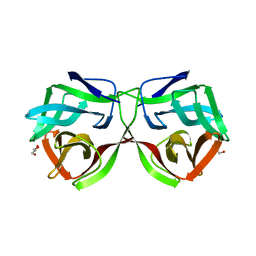 | |
6ZY3
 
 | | Cryo-EM structure of MlaFEDB in complex with phospholipid | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ... | | 著者 | Dong, C.J, Dong, H.H. | | 登録日 | 2020-07-30 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZY2
 
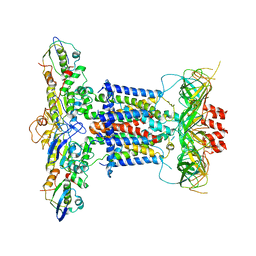 | | Cryo-EM structure of apo MlaFEDB | | 分子名称: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, Toluene tolerance protein Ttg2A, ... | | 著者 | Dong, C.J, Dong, H.H. | | 登録日 | 2020-07-30 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZY4
 
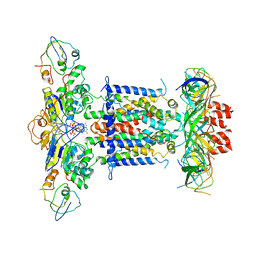 | | Cryo-EM structure of MlaFEDB in complex with ADP | | 分子名称: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Dong, C.J, Dong, H.H. | | 登録日 | 2020-07-30 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZY9
 
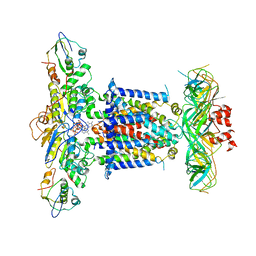 | | Cryo-EM structure of MlaFEDB in complex with AMP-PNP | | 分子名称: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, MAGNESIUM ION, ... | | 著者 | Dong, C.J, Dong, H.H. | | 登録日 | 2020-07-30 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5MH2
 
 | |
5MH0
 
 | |
