8GPG
 
 | | HIV-1 Env X18 UFO in complex with F6 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, F6 Fab heavy chain, ... | | 著者 | Niu, J, Xu, Y.W, Yang, B. | | 登録日 | 2022-08-26 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structures and immune recognition of Env trimers from two Asia prevalent HIV-1 CRFs.
Nat Commun, 14, 2023
|
|
6V63
 
 | | SETD3 WT in Complex with an Actin Peptide with His73 Replaced with Glutamine | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Actin, ... | | 著者 | Dai, S, Horton, J.R, Cheng, X. | | 登録日 | 2019-12-04 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | An engineered variant of SETD3 methyltransferase alters target specificity from histidine to lysine methylation.
J.Biol.Chem., 295, 2020
|
|
5D6P
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[4-(hydroxymethyl)-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl]urea, DNA gyrase subunit B, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-12 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
8D4K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4M
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4N
 
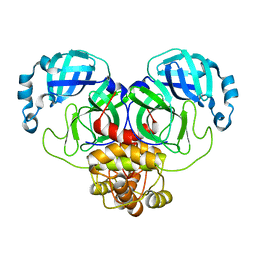 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166Q Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4J
 
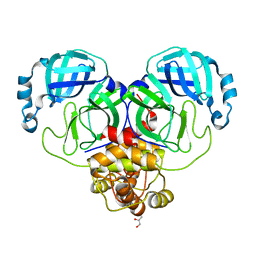 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant | | 分子名称: | 3C-like proteinase nsp5, GLYCEROL | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
5K5J
 
 | |
5D6Q
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-{4-[(E)-2-(pyridin-3-yl)ethenyl]-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl}urea, DNA gyrase subunit B, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-12 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5KE9
 
 | |
5KEA
 
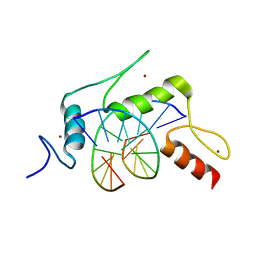 | |
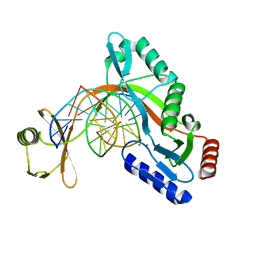
6UKH
 
 | |
7RTB
 
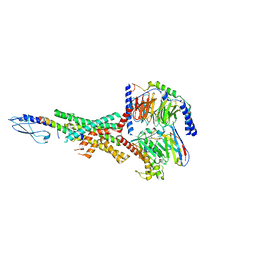 | | Peptide-19 bound to the Glucagon-Like Peptide-1 Receptor (GLP-1R) | | 分子名称: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Johnson, R.M, Danev, R, Sexton, P.M, Wootten, D. | | 登録日 | 2021-08-12 | | 公開日 | 2021-10-06 | | 実験手法 | ELECTRON MICROSCOPY (2.14 Å) | | 主引用文献 | Cryo-EM structure of the dual incretin receptor agonist, peptide-19, in complex with the glucagon-like peptide-1 receptor.
Biochem.Biophys.Res.Commun., 578, 2021
|
|
5KL6
 
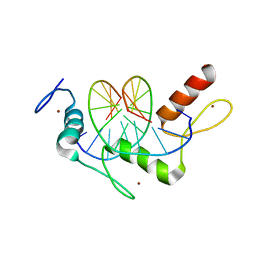 | | Wilms Tumor Protein (WT1) Q369R ZnF2-4 in complex with DNA | | 分子名称: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*GP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*CP*CP*CP*CP*AP*CP*GP*C)-3'), Wilms tumor protein, ... | | 著者 | Hashimoto, H, Cheng, X. | | 登録日 | 2016-06-23 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.641 Å) | | 主引用文献 | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
7S0V
 
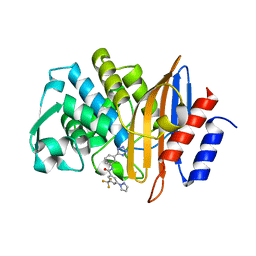 | |
5KL4
 
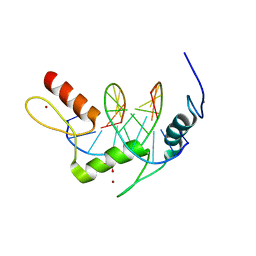 | |
5KKQ
 
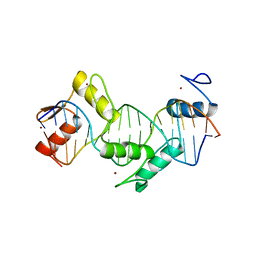 | | Homo sapiens CCCTC-binding factor (CTCF) ZnF3-7 and DNA complex structure | | 分子名称: | DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), Transcriptional repressor CTCF, ... | | 著者 | Hashimoto, H, Wang, D, Cheng, X. | | 登録日 | 2016-06-22 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.744 Å) | | 主引用文献 | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
5KL5
 
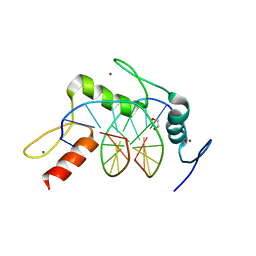 | | Wilms Tumor Protein (WT1) ZnF2-4 Q369H in complex with carboxylated DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(1CC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | 著者 | Hashimoto, H, Cheng, X. | | 登録日 | 2016-06-23 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.289 Å) | | 主引用文献 | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
5D7C
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[1-(pyridin-2-yl)-6-(pyridin-3-yl)-1H-pyrrolo[3,2-b]pyridin-3-yl]urea, DNA gyrase subunit B, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-13 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5KL2
 
 | | Wilms Tumor Protein (WT1) ZnF2-4 in complex with DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | 著者 | Hashimoto, H, Cheng, X. | | 登録日 | 2016-06-23 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.692 Å) | | 主引用文献 | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
5KE7
 
 | |
7VQ1
 
 | | Structure of Apo-hsTRPM2 channel | | 分子名称: | Transient receptor potential cation channel subfamily M member 2 | | 著者 | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | 登録日 | 2021-10-18 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
7VQ2
 
 | | Structure of Apo-hsTRPM2 channel TM domain | | 分子名称: | Transient receptor potential cation channel subfamily M member 2 | | 著者 | Yu, X.F, Xie, Y, Zhang, X.K, Ma, C, Guo, J.T, Yang, F, Yang, W. | | 登録日 | 2021-10-18 | | 公開日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | Structural and functional basis of the selectivity filter as a gate in human TRPM2 channel.
Cell Rep, 37, 2021
|
|
7SL2
 
 | |
7SL6
 
 | |
