3LI1
 
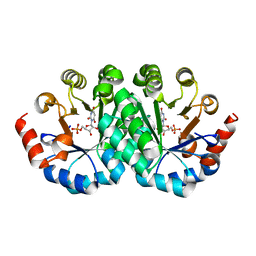 | | Crystal structure of the mutant I218A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-01-23 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LHT
 
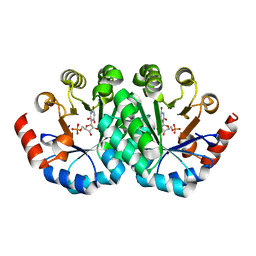 | | Crystal structure of the mutant V201F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-01-23 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LLD
 
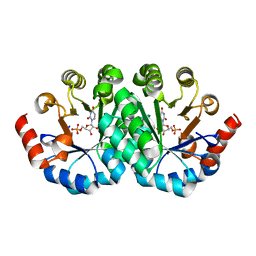 | | Crystal structure of the mutant S127G of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | 分子名称: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-01-28 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
5HEL
 
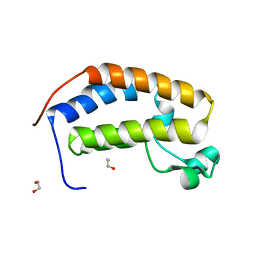 | | Crystal structure of the N-terminus Y153H bromodomain mutant of human BRD2 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2 | | 著者 | Tallant, C, Lori, C, Pasquo, A, Chiaraluce, R, Consalvi, V, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | 登録日 | 2016-01-06 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal structure of the N-terminus Y153H bromodomain mutant of human BRD2
To Be Published
|
|
3LOB
 
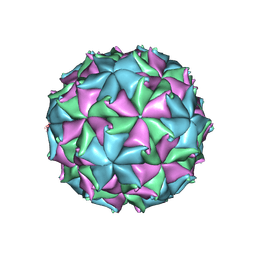 | | Crystal Structure of Flock House Virus calcium mutant | | 分子名称: | Coat protein beta, Coat protein gamma, RNA (5'-R(*UP*UP*U*AP*UP*CP*UP*(P))-3'), ... | | 著者 | Johnson, J.E, Banerjee, M, Speir, J.A, Huang, R. | | 登録日 | 2010-02-03 | | 公開日 | 2010-04-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structure and function of a genetically engineered mimic of a nonenveloped virus entry intermediate.
J.Virol., 84, 2010
|
|
5E7M
 
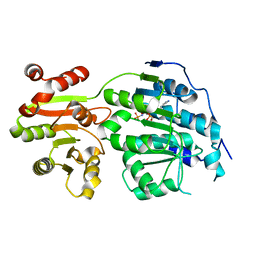 | |
2BAY
 
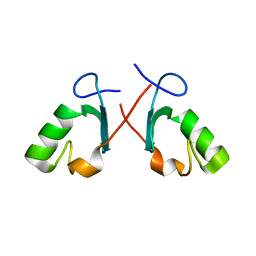 | | Crystal structure of the Prp19 U-box dimer | | 分子名称: | Pre-mRNA splicing factor PRP19 | | 著者 | Vander Kooi, C.W, Ohi, M.D, Rosenberg, J.A, Oldham, M.L, Newcomer, M.E, Gould, K.L, Chazin, W.J. | | 登録日 | 2005-10-15 | | 公開日 | 2006-01-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The Prp19 U-box Crystal Structure Suggests a Common Dimeric Architecture for a Class of Oligomeric E3 Ubiquitin Ligases.
Biochemistry, 45, 2006
|
|
3LHU
 
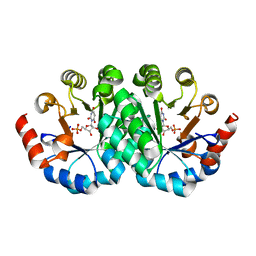 | | Crystal structure of the mutant I199F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-01-23 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LLF
 
 | | Crystal structure of the mutant S127P of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | 分子名称: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-01-29 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
5DS6
 
 | | Crystal structure the Escherichia coli Cas1-Cas2 complex bound to protospacer DNA with splayed ends | | 分子名称: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | 著者 | Nunez, J.K, Harrington, L.B, Kranzusch, P.J, Engelman, A.N, Doudna, J.A. | | 登録日 | 2015-09-16 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.352 Å) | | 主引用文献 | Foreign DNA capture during CRISPR-Cas adaptive immunity.
Nature, 527, 2015
|
|
5DXR
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-{[(3R)-3-methylcyclohexylidene]methanediyl}diphenol | | 分子名称: | 4,4'-{[(3R)-3-methylcyclohexylidene]methanediyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-23 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
2AV8
 
 | |
5HEN
 
 | | Crystal structure of the N-terminus R100L bromodomain mutant of human BRD2 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2 | | 著者 | Tallant, C, Lori, C, Pasquo, A, Chiaraluce, R, Consalvi, V, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | 登録日 | 2016-01-06 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Crystal structure of the N-terminus R100L bromodomain mutant of human BRD2
To Be Published
|
|
5H9A
 
 | | Crystal structure of the Apo form of human cellular retinol binding protein 1 | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Retinol-binding protein 1 | | 著者 | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | 登録日 | 2015-12-26 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.381 Å) | | 主引用文献 | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
2CW6
 
 | | Crystal Structure of Human HMG-CoA Lyase: Insights into Catalysis and the Molecular Basis for Hydroxymethylglutaric Aciduria | | 分子名称: | 3-HYDROXYPENTANEDIOIC ACID, Hydroxymethylglutaryl-CoA lyase, mitochondrial, ... | | 著者 | Fu, Z, Runquist, J.A, Hunt, J.F, Miziorko, H.M, Kim, J.-J.P. | | 登録日 | 2005-06-17 | | 公開日 | 2005-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of human 3-hydroxy-3-methylglutaryl-CoA Lyase: insights into catalysis and the molecular basis for hydroxymethylglutaric aciduria
J.Biol.Chem., 281, 2006
|
|
1T38
 
 | | HUMAN O6-ALKYLGUANINE-DNA ALKYLTRANSFERASE BOUND TO DNA CONTAINING O6-METHYLGUANINE | | 分子名称: | 5'-D(*GP*CP*CP*AP*TP*GP*(6OG)P*CP*TP*AP*GP*TP*A)-3', 5'-D(*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*GP*C)-3', Methylated-DNA--protein-cysteine methyltransferase | | 著者 | Daniels, D.S, Woo, T.T, Luu, K.X, Noll, D.M, Clarke, N.D, Pegg, A.E, Tainer, J.A. | | 登録日 | 2004-04-25 | | 公開日 | 2004-07-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | DNA binding and nucleotide flipping by the human DNA repair protein AGT.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3HPF
 
 | | Crystal structure of the mutant Y90F of divergent galactarate dehydratase from Oceanobacillus iheyensis complexed with Mg and galactarate | | 分子名称: | D-galactaric acid, MAGNESIUM ION, Muconate cycloisomerase | | 著者 | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Gerlt, J.A, Almo, S.C. | | 登録日 | 2009-06-04 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
3I4K
 
 | | Crystal structure of Muconate lactonizing enzyme from Corynebacterium glutamicum | | 分子名称: | ACETIC ACID, MAGNESIUM ION, Muconate lactonizing enzyme | | 著者 | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-07-01 | | 公開日 | 2009-07-14 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of Muconate lactonizing enzyme from Corynebacterium glutamicum
To be Published
|
|
2FD3
 
 | |
3HUE
 
 | |
2FLA
 
 | | Structure of thereduced HiPIP from thermochromatium tepidum at 0.95 angstrom resolution | | 分子名称: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | 著者 | Hunsicker-Wang, L.M, Stout, C.D, Fee, J.A. | | 登録日 | 2006-01-05 | | 公開日 | 2006-12-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Geometric factors determine, in part, the electronic state of the 4Fe-4S cluster of HiPIP: a geometric, crystallographic, and theoretical study.
To be Published
|
|
5HQJ
 
 | | Crystal structure of ABC transporter Solute Binding Protein B1G1H7 from Burkholderia graminis C4D1M, target EFI-511179, in complex with D-arabinose | | 分子名称: | CHLORIDE ION, Periplasmic binding protein/LacI transcriptional regulator, alpha-D-arabinopyranose | | 著者 | Roth, Y, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2016-01-21 | | 公開日 | 2016-03-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of ABC transporter Solute Binding Protein B1G1H7 from Burkholderia graminis C4D1M, target EFI-511179, in complex with D-arabinose
To be published
|
|
3LK4
 
 | | Crystal structure of CapZ bound to the uncapping motif from CD2AP | | 分子名称: | CD2-associated protein, F-actin-capping protein subunit alpha-1, F-actin-capping protein subunit beta isoforms 1 and 2 | | 著者 | Hernandez-Valladares, M, Kim, T, Kannan, B, Tung, A, Cooper, J.A, Robinson, R.C. | | 登録日 | 2010-01-27 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural characterization of a capping protein interaction motif defines a family of actin filament regulators.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5HUR
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25T/I92K at cryogenic temperature | | 分子名称: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | 著者 | Skerritt, L.A, Caro, J.A, Heroux, A, Schlessman, J.L, Garcia-Moreno E, B. | | 登録日 | 2016-01-27 | | 公開日 | 2016-02-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25T/I92K at cryogenic temperature
To be Published
|
|
3LHW
 
 | | Crystal structure of the mutant V182A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-01-23 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
