7C8V
 
 | | Structure of sybody SR4 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | 著者 | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | 登録日 | 2020-06-03 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7C8W
 
 | | Structure of sybody MR17 in complex with the SARS-CoV-2 S receptor-binding domain (RBD) | | 分子名称: | GLYCEROL, Spike protein S1, Synthetic nanobody MR17, ... | | 著者 | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | 登録日 | 2020-06-03 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
8X55
 
 | | BA.2.86 Spike Trimer with T356K mutation (3 RBD down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
6NZV
 
 | | Crystal structure of HCV NS3/4A protease in complex with compound 12 | | 分子名称: | (1aR,5S,8S,9S,10R,22aR)-5-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-9-ethyl-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, HCV NS3/4A protease, SULFATE ION, ... | | 著者 | Appleby, T.C, Taylor, J.G. | | 登録日 | 2019-02-14 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Discovery of the pan-genotypic hepatitis C virus NS3/4A protease inhibitor voxilaprevir (GS-9857): A component of Vosevi®.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
8X4Z
 
 | | BA.2.86 Spike Trimer with ins483V mutation (3 RBD down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X56
 
 | | BA.2.86 Spike Trimer with T356K mutation (1 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X50
 
 | | BA.2.86 Spike Trimer with ins483V mutation (1 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.82 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X4H
 
 | | SARS-CoV-2 JN.1 Spike | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-15 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X5R
 
 | | SARS-CoV-2 BA.2.75 Spike with K356T mutation (1 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-17 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUR
 
 | | BA.2.86 Spike Trimer in complex with heparan sulfate | | 分子名称: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yue, C, Liu, P. | | 登録日 | 2024-01-14 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUT
 
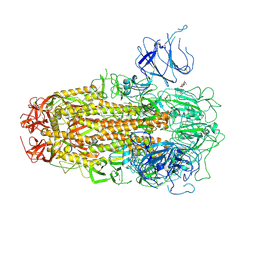 | | XBB.1.5 Spike Trimer in complex with heparan sulfate | | 分子名称: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yue, C, Liu, P, Mao, X. | | 登録日 | 2024-01-14 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUU
 
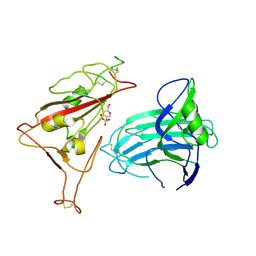 | |
8XUS
 
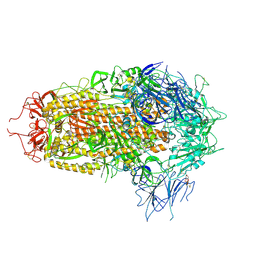 | | JN.1 Spike Trimer in complex with heparan sulfate | | 分子名称: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yue, C, Liu, P. | | 登録日 | 2024-01-14 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X5Q
 
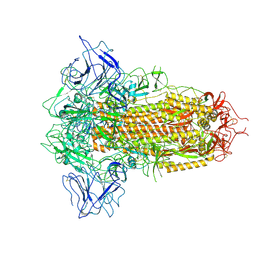 | | SARS-CoV-2 BA.2.75 Spike with K356T mutation (3 RBD down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-17 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
4LN0
 
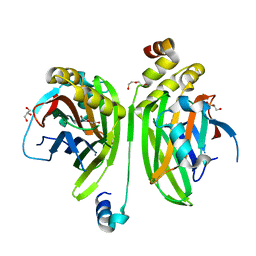 | | Crystal structure of the VGLL4-TEAD4 complex | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Transcription cofactor vestigial-like protein 4, ... | | 著者 | Wang, H, Shi, Z, Zhou, Z. | | 登録日 | 2013-07-11 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.896 Å) | | 主引用文献 | A Peptide Mimicking VGLL4 Function Acts as a YAP Antagonist Therapy against Gastric Cancer.
Cancer Cell, 25, 2014
|
|
7CWS
 
 | |
7CAN
 
 | | Structure of sybody MR17-K99Y in complex with the SARS-CoV-2 S Receptor-binding domain (RBD) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | 著者 | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | 登録日 | 2020-06-09 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
5WB6
 
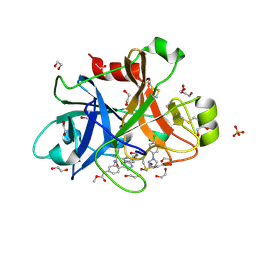 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(11S)-11-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-6-fluoro-2-oxo-1,3,4,10,11,13-hexahydro-2H-5,9:15,12-di(azeno)-1,13-benzodiazacycloheptadecin-18-yl]carbamate | | 分子名称: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | 著者 | Sheriff, S. | | 登録日 | 2017-06-28 | | 公開日 | 2017-08-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Macrocyclic factor XIa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7CWU
 
 | |
3IK3
 
 | |
5H25
 
 | | EED in complex with PRC2 allosteric inhibitor compound 11 | | 分子名称: | 5-(2-fluorophenyl)-2,3-dihydroimidazo[2,1-a]isoquinoline, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | 著者 | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | 登録日 | 2016-10-14 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
6BRR
 
 | |
4OCH
 
 | | Apo structure of Smr domain of MutS2 from Deinococcus radiodurans | | 分子名称: | Endonuclease MutS2, GLYCEROL | | 著者 | Zhang, H, Zhao, Y, Xu, Q, Hua, Y.J. | | 登録日 | 2014-01-09 | | 公開日 | 2014-06-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.4001 Å) | | 主引用文献 | Structural and functional studies of MutS2 from Deinococcus radiodurans.
Dna Repair, 21, 2014
|
|
5EOA
 
 | | Crystal structure of OPTN E50K mutant and TBK1 complex | | 分子名称: | Optineurin, Serine/threonine-protein kinase TBK1 | | 著者 | Li, F, Xie, X, Liu, J, Pan, L. | | 登録日 | 2015-11-10 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.503 Å) | | 主引用文献 | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
5EOB
 
 | | Crystal structure of CMET in complex with novel inhibitor | | 分子名称: | 6-[bis(fluoranyl)-[6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl]methyl]quinoline, Hepatocyte growth factor receptor | | 著者 | Liu, Q, Chen, T, Xu, Y. | | 登録日 | 2015-11-10 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Discovery of 6-(difluoro(6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl)methyl)quinoline as a highly potent and selective c-Met inhibitor
Eur.J.Med.Chem., 116, 2016
|
|
