7ERB
 
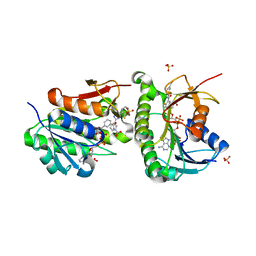 | | Crystal structure of human Biliverdin IX-beta reductase B with Ataluren (PTC) | | 分子名称: | 3-[5-(2-fluorophenyl)-1,2,4-oxadiazol-3-yl]benzoic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | 登録日 | 2021-05-06 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
9L8R
 
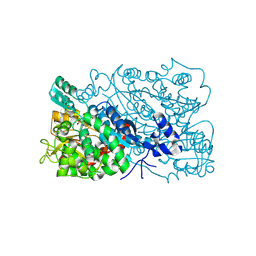 | | Dihydroxyacid dehydratase (DHAD) | | 分子名称: | (2~{R})-7-fluoranyl-2-oxidanyl-3-oxidanylidene-4-prop-2-ynyl-1,4-benzoxazine-2-carboxylic acid, Dihydroxy-acid dehydratase, chloroplastic, ... | | 著者 | Zhou, J, Liu, D. | | 登録日 | 2024-12-28 | | 公開日 | 2025-06-04 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | The mutant structure of DHAD
To Be Published
|
|
9LR5
 
 | |
9M0S
 
 | |
7ERA
 
 | | Crystal structure of human Biliverdin IX-beta reductase B with Olsalazine Sodium (OSS) | | 分子名称: | 5-[(E)-(3-carboxy-4-oxidanyl-phenyl)diazenyl]-2-oxidanyl-benzoic acid, Flavin reductase (NADPH), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Griesinger, C, Lee, D, Ryu, K.S, Kim, M, Ha, J.H. | | 登録日 | 2021-05-06 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Repositioning Food and Drug Administration-Approved Drugs for Inhibiting Biliverdin IX beta Reductase B as a Novel Thrombocytopenia Therapeutic Target.
J.Med.Chem., 65, 2022
|
|
7FC9
 
 | | Crystal structure of CmABCB1 in lipidic mesophase revealed by LCP-SFX | | 分子名称: | ACETATE ION, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Pan, D, Oyama, R, Sato, T, Nakane, T, Mizunuma, R, Matsuoka, K, Joti, Y, Tono, K, Nango, E, Iwata, S, Nakatsu, T, Kato, H. | | 登録日 | 2021-07-14 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of CmABCB1 multi-drug exporter in lipidic mesophase revealed by LCP-SFX.
Iucrj, 9, 2022
|
|
9L99
 
 | | Structure of Western equine encephalitis virus McMillan strain in complex with VLDLR LA1-2 | | 分子名称: | CALCIUM ION, Spike glycoprotein E1, Spike glycoprotein E2, ... | | 著者 | Ma, B, Cao, Z, Ding, W, Zhang, X, Xiang, Y, Cao, D. | | 登録日 | 2024-12-29 | | 公開日 | 2025-06-04 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis for the recognition of two different types of receptors by Western equine encephalitis virus.
Cell Rep, 44, 2025
|
|
9R2Q
 
 | | p53 bound to nucleosome at position SHL-5.7 (non-crosslinked sample) | | 分子名称: | Cellular tumor antigen p53, DNA-for, DNA-rev, ... | | 著者 | Chakraborty, D, Michael, A.K, Kempf, G, Cavadini, S, Kater, L, Thoma, N.H. | | 登録日 | 2025-04-30 | | 公開日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | p53 bound to nucleosome at position SHL-5.7 (non-crosslinked sample)
To Be Published
|
|
9L9A
 
 | | Structure of Western equine encephalitis virus McMillan strain in complex with VLDLR LA2-3 | | 分子名称: | CALCIUM ION, Spike glycoprotein E1, Spike glycoprotein E2, ... | | 著者 | Ma, B, Cao, Z, Ding, W, Zhang, X, Xiang, Y, Cao, D. | | 登録日 | 2024-12-29 | | 公開日 | 2025-06-04 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis for the recognition of two different types of receptors by Western equine encephalitis virus.
Cell Rep, 44, 2025
|
|
9QQ6
 
 | | Structure of the Azotobacter vinelandii NifL-NifA complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Nif-specific regulatory protein, ... | | 著者 | Bueno Batista, M, Richardson, J, Webster, M.W, Ghilarov, D, Peters, J.W, Lawson, D.M, Dixon, R. | | 登録日 | 2025-03-31 | | 公開日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (6.45 Å) | | 主引用文献 | Structural and functional analysis of the NifL-NifA complex for engineered control of nitrogen fixation in Proteobacteria
To Be Published
|
|
9NIU
 
 | | Co-crystal structure of FABP7 in complex with PFO3TDA | | 分子名称: | 1,2-ETHANEDIOL, Fatty acid-binding protein, brain, ... | | 著者 | Yang, D, Liu, J, Zeng, H, Dong, A, Arrowsmith, C.H, Edwards, A.M, Peng, H, Halabelian, L, Structural Genomics Consortium (SGC) | | 登録日 | 2025-02-26 | | 公開日 | 2025-05-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Co-crystal structure of FABP7 in complex with PFO3TDA
To be published
|
|
9NWC
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-276-5Br | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(5-bromo-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-D-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Bulut, H, Kuwata, N, Hayashi, H, Aoki, H, Hattori, S, Li, M, Das, D, Misumi, S, Wlodawer, A, Tamamura, H, Mitsuya, H. | | 登録日 | 2025-03-21 | | 公開日 | 2025-06-04 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Impact of Single Halogen Atom Substitutions on Potency of Inhibitors Targeting SARS-CoV-2 Main Protease
To Be Published
|
|
9R04
 
 | | p53 bound to the nucleosome at position SHL-5.7 (crosslinked sample) | | 分子名称: | Cellular tumor antigen p53, DNA-for, DNA-rev, ... | | 著者 | Chakraborty, D, Michael, A.K, Kempf, G, Cavadini, S, Kater, L, Thoma, N.H. | | 登録日 | 2025-04-24 | | 公開日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | p53 bound to the nucleosome at position SHL-5.7 (crosslinked sample)
To Be Published
|
|
9R2M
 
 | | p53 bound to nucleosome at position SHL+5.9 (non-crosslinked sample, composite map) | | 分子名称: | Cellular tumor antigen p53, DNA-for, DNA-rev, ... | | 著者 | Chakraborty, D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2025-04-30 | | 公開日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | p53 bound to nucleosome at position SHL+5.9 (non-crosslinked sample, composite map)
To Be Published
|
|
9QCG
 
 | |
9R6Y
 
 | |
9R70
 
 | |
9MUJ
 
 | | RlmR 23S rRNA methyltransferase from Thermus thermophilus in complex with methylated rRNA (Um2552) and S-adenosyl-L-homocysteine (SAH) | | 分子名称: | 23S rRNA methyltransferase, RNA (59-MER), S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Tanouti, Y, Roovers, M, Droogmans, L, Van Elder, D, Kruys, V, Labar, G. | | 登録日 | 2025-01-14 | | 公開日 | 2025-04-23 | | 最終更新日 | 2025-06-18 | | 実験手法 | X-RAY DIFFRACTION (2.006 Å) | | 主引用文献 | Structural insight into the novel Thermus thermophilus SPOUT methyltransferase RlmR catalysing Um2552 formation in the 23S rRNA A-loop: a case of convergent evolution.
Nucleic Acids Res., 53, 2025
|
|
9NLP
 
 | | HIV-1 Reverse Transcriptase with New Non-Nucleoside Reverse Transcriptase Inhibitor 12126065 | | 分子名称: | 4-({5-amino-1-[6-(2-cyanoethyl)naphthalene-1-sulfonyl]-1H-1,2,4-triazol-3-yl}amino)-2-chlorobenzonitrile, Reverse transcriptase p51 subunit, Reverse transcriptase p66 subunit | | 著者 | Young, M.A, Lane, T.R, Raman, R, Nelson, J.A.E, Riabova, O, Kazakova, E, Monakhova, N, Tsedilin, A, Rees, S.D, Quinnell, D, Chang, G, Ekins, S. | | 登録日 | 2025-03-03 | | 公開日 | 2025-05-07 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | Cryo-EM Structure of HIV-1 Reverse Transcriptase with N -Phenyl-1-(phenylsulfonyl)-1 H -1,2,4-triazol-3-amine: A New HIV-1 Non-nucleoside Inhibitor.
Acs Infect Dis., 11, 2025
|
|
9LSL
 
 | | The crystal structure of PDE5A with L1 | | 分子名称: | (13~{S},15~{R})-15-(3-chloranyl-4-methoxy-phenyl)-12-ethanoyl-8,12,16-triazatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(17),2,4,6,8-pentaene-4-carbonitrile, MAGNESIUM ION, ZINC ION, ... | | 著者 | Wu, D, Huang, Y.-Y, Luo, H.-B. | | 登録日 | 2025-02-04 | | 公開日 | 2025-06-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Plasma metabolites-based drug design: Discovery of novel and highly selective phosphodiesterase 5 inhibitors
Chin.Chem.Lett., 2025
|
|
9M2Q
 
 | | Imidazole glycerol phosphate dehydratase from Mycobacterium tuberculosis, in complex with aminotriazole | | 分子名称: | 3-AMINO-1,2,4-TRIAZOLE, Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION | | 著者 | Raina, R, Kar, D, Singla, M, Tiwari, S, Kumari, S, Aneja, S, Kumar, V, Banerjee, S, Goyal, S, Pal, R.K, Vinothkumar, K.R, Biswal, B.K. | | 登録日 | 2025-02-28 | | 公開日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Cryo-EM structures of Mycobacterium tuberculosis imidazole glycerol phosphate dehydratase in the apo state and in the presence of small molecules.
Acta Crystallogr.,Sect.F, 2025
|
|
9QFV
 
 | | Human Tryptase beta-2 (hTPSB2) | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | 著者 | Porta, A, Manelfi, C, Talarico, C, Beccari, A.R, Brindisi, M, Summa, V, Iaconis, D, Gobbi, M, Beeg, M. | | 登録日 | 2025-03-12 | | 公開日 | 2025-03-26 | | 最終更新日 | 2025-04-09 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Integrating Surface Plasmon Resonance and Docking Analysis for Mechanistic Insights of Tryptase Inhibitors.
Molecules, 30, 2025
|
|
7F0S
 
 | |
7FIO
 
 | | LecA from Pseudomonas aeruginosa in complex with a synthetic monovalent galactosidic inhibitor | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, N-[2-[4-[(2S)-3-(2-hydroxyethylamino)-3-oxidanylidene-2-(2-phenoxyethanoylamino)propyl]-1,2,3-triazol-1-yl]ethyl]-4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-benzamide, ... | | 著者 | Kuhaudomlarp, S, Siebs, E, da Silva Figueiredo Celestino Gomes, P, Fortin, C, Rognan, D, Rademacher, C, Imberty, A, Titz, A. | | 登録日 | 2021-07-31 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Targeting the Central Pocket of the Pseudomonas aeruginosa Lectin LecA.
Chembiochem, 23, 2022
|
|
9QWO
 
 | | Vinculin tail bound to paxillin LD2 | | 分子名称: | ACETATE ION, Isoform 1 of Vinculin, Isoform Gamma of Paxillin, ... | | 著者 | Diaz-Palacios, K, Lietha, D. | | 登録日 | 2025-04-14 | | 公開日 | 2025-04-23 | | 最終更新日 | 2025-05-07 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Phospho-regulated tethering of focal adhesion kinase to vinculin links force transduction to focal adhesion signaling.
Cell Commun Signal, 23, 2025
|
|
