8HWN
 
 | |
7WMD
 
 | |
7WMK
 
 | | PQQ-dependent alcohol dehydrogenase complexed with PQQ | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | 著者 | Chen, M, Yang, H, Lv, F. | | 登録日 | 2022-01-15 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Structure-Function Analysis of a Quinone-Dependent Dehydrogenase Capable of Deoxynivalenol Detoxification.
J.Agric.Food Chem., 70, 2022
|
|
3PHD
 
 | | Crystal structure of human HDAC6 in complex with ubiquitin | | 分子名称: | Histone deacetylase 6, Polyubiquitin, ZINC ION | | 著者 | Dong, A, Qui, W, Ravichandran, M, Schuetz, A, Loppnau, P, Li, F, Mackenzie, F, Kozieradzki, I, Ouyang, H, Structural Genomics Consortium (SGC) | | 登録日 | 2010-11-03 | | 公開日 | 2011-02-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Protein Aggregates Are Recruited to Aggresome by Histone Deacetylase 6 via Unanchored Ubiquitin C Termini.
J.Biol.Chem., 287, 2012
|
|
4UBD
 
 | | Crystal structure of a neutralizing human monoclonal antibody with 1968 H3 HA | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | 著者 | Shore, D.A, Yang, H, Cho, M, Donis, R.O, Stevens, J. | | 登録日 | 2014-08-12 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus.
Nat Commun, 6, 2015
|
|
3DSF
 
 | | Crystal structure of anti-osteopontin antibody 23C3 in complex with W43A mutated epitope peptide | | 分子名称: | Fab fragment of anti-osteopontin antibody 23C3, Heavy chain, Light chain, ... | | 著者 | Du, J, Zhong, C, Yang, H, Ding, J. | | 登録日 | 2008-07-12 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Molecular basis of recognition of human osteopontin by 23C3, a potential therapeutic antibody for treatment of rheumatoid arthritis
J.Mol.Biol., 382, 2008
|
|
3CMT
 
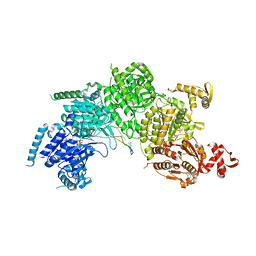 | | Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*DTP*DTP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DCP*DTP*DTP*DTP*DT)-3'), DNA (5'-D(P*DGP*DGP*DTP*DGP*DGP*DG)-3'), ... | | 著者 | Chen, Z, Yang, H, Pavletich, N.P. | | 登録日 | 2008-03-24 | | 公開日 | 2008-05-20 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures.
Nature, 453, 2008
|
|
4KTH
 
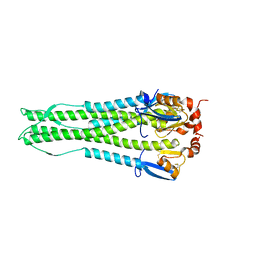 | | Structure of A/Hubei/1/2010 H5 HA | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | 著者 | Shore, D.A, Yang, H, Carney, P.J, Chang, J.C, Stevens, J. | | 登録日 | 2013-05-20 | | 公開日 | 2013-11-27 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural and Antigenic Variation among Diverse Clade 2 H5N1 Viruses.
Plos One, 8, 2013
|
|
3JZN
 
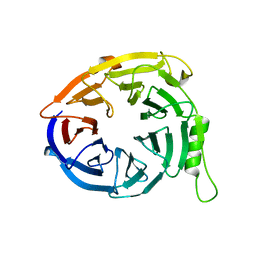 | | Structure of EED in apo form | | 分子名称: | Polycomb protein EED | | 著者 | Xu, C, Bian, C.B, Ouyang, H, Qiu, W, MacKenzie, F, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2009-09-23 | | 公開日 | 2009-12-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1D5R
 
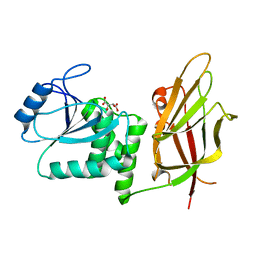 | | Crystal Structure of the PTEN Tumor Suppressor | | 分子名称: | L(+)-TARTARIC ACID, PHOSPHOINOSITIDE PHOSPHATASE PTEN | | 著者 | Lee, J.O, Yang, H, Georgescu, M.-M, Di Cristofano, A, Pavletich, N.P. | | 登録日 | 1999-10-11 | | 公開日 | 1999-11-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association.
Cell(Cambridge,Mass.), 99, 1999
|
|
1DK4
 
 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE | | 分子名称: | INOSITOL MONOPHOSPHATASE, PHOSPHATE ION, ZINC ION | | 著者 | Stec, B, Yang, H, Johnson, K.A, Chen, L, Roberts, M.F. | | 登録日 | 1999-12-06 | | 公開日 | 2000-11-08 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | MJ0109 is an enzyme that is both an inositol monophosphatase and the 'missing' archaeal fructose-1,6-bisphosphatase.
Nat.Struct.Biol., 7, 2000
|
|
1LBV
 
 | | Crystal Structure of apo-form (P21) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus | | 分子名称: | fructose 1,6-bisphosphatase/inositol monophosphatase | | 著者 | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | 登録日 | 2002-04-04 | | 公開日 | 2002-05-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
1LBX
 
 | | Crystal Structure of a ternary complex of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus with Calcium ions and D-myo-Inositol-1-Phosphate | | 分子名称: | CALCIUM ION, D-MYO-INOSITOL-1-PHOSPHATE, fructose 1,6-bisphosphatase/inositol monophosphatase | | 著者 | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | 登録日 | 2002-04-04 | | 公開日 | 2002-05-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
1LBZ
 
 | | Crystal Structure of a complex (P32 crystal form) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus with 3 Calcium ions and Fructose-1,6 bisphosphate | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, CALCIUM ION, fructose 1,6-bisphosphatase/inositol monophosphatase | | 著者 | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | 登録日 | 2002-04-04 | | 公開日 | 2002-05-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
1LBY
 
 | | Crystal Structure of a complex (P32 crystal form) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus with 3 Manganese ions, Fructose-6-Phosphate, and Phosphate ion | | 分子名称: | 6-O-phosphono-beta-D-fructofuranose, MANGANESE (II) ION, PHOSPHATE ION, ... | | 著者 | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | 登録日 | 2002-04-04 | | 公開日 | 2002-05-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
1LBW
 
 | | Crystal Structure of apo-form (P32) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus | | 分子名称: | fructose 1,6-bisphosphatase/inositol monophosphatase | | 著者 | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | 登録日 | 2002-04-04 | | 公開日 | 2002-05-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
7A5K
 
 | | Structure of the human mitoribosome in the post translocation state bound to mtEF-G1 | | 分子名称: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | 著者 | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | 登録日 | 2020-08-21 | | 公開日 | 2020-12-23 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7A5J
 
 | | Structure of the split human mitoribosomal large subunit with P-and E-site mt-tRNAs | | 分子名称: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | 著者 | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | 登録日 | 2020-08-21 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7A5I
 
 | | Structure of the human mitoribosome with A- P-and E-site mt-tRNAs | | 分子名称: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | 著者 | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | 登録日 | 2020-08-21 | | 公開日 | 2020-12-23 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7A5G
 
 | | Structure of the elongating human mitoribosome bound to mtEF-Tu.GMPPCP and A/T mt-tRNA | | 分子名称: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | 著者 | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | 登録日 | 2020-08-21 | | 公開日 | 2020-12-23 | | 実験手法 | ELECTRON MICROSCOPY (4.33 Å) | | 主引用文献 | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7A5H
 
 | | Structure of the split human mitoribosomal large subunit with rescue factors mtRF-R and MTRES1 | | 分子名称: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | 著者 | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | 登録日 | 2020-08-21 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
7A5F
 
 | | Structure of the stalled human mitoribosome with P- and E-site mt-tRNAs | | 分子名称: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | 著者 | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | 登録日 | 2020-08-21 | | 公開日 | 2020-12-23 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
1NH9
 
 | | Crystal Structure of a DNA Binding Protein Mja10b from the hyperthermophile Methanococcus jannaschii | | 分子名称: | DNA-binding protein Alba | | 著者 | Wang, G, Bartlam, M, Guo, R, Yang, H, Xue, H, Liu, Y, Huang, L, Rao, Z. | | 登録日 | 2002-12-19 | | 公開日 | 2003-12-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a DNA binding protein from the hyperthermophilic euryarchaeon Methanococcus jannaschii
Protein Sci., 12, 2003
|
|
8PUF
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 20 degrees | | 分子名称: | Gag protein (Fragment) | | 著者 | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | 登録日 | 2023-07-17 | | 公開日 | 2023-08-23 | | 最終更新日 | 2024-09-18 | | 実験手法 | ELECTRON MICROSCOPY (6.1 Å) | | 主引用文献 | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 2024
|
|
8PUG
 
 | | Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-NTD refinement | | 分子名称: | Gag polyprotein | | 著者 | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | 登録日 | 2023-07-17 | | 公開日 | 2023-08-23 | | 最終更新日 | 2024-09-18 | | 実験手法 | ELECTRON MICROSCOPY (5.9 Å) | | 主引用文献 | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 2024
|
|
