6KC6
 
 | | HOIP-HOIPIN8 complex | | 分子名称: | 2-[3-[2,6-bis(fluoranyl)-4-(1~{H}-pyrazol-4-yl)phenyl]-3-oxidanylidene-propyl]-4-(1-methylpyrazol-4-yl)benzoic acid, CHLORIDE ION, E3 ubiquitin-protein ligase RNF31, ... | | 著者 | Sato, Y, Fukai, S. | | 登録日 | 2019-06-27 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.123 Å) | | 主引用文献 | Molecular bases for HOIPINs-mediated inhibition of LUBAC and innate immune responses.
Commun Biol, 3, 2020
|
|
6K36
 
 | |
6K37
 
 | | Crystal structure of BioU (K124A) from Synechocystis sp.PCC6803 in complex with NAD+ and the analog of reaction intermediate, 3-(1-aminoethyl)-nonanedioic acid | | 分子名称: | (3R)-3-[(1R)-1-azanylethyl]nonanedioic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Slr0355 protein | | 著者 | Sakaki, K, Tomita, T, Nishiyama, M. | | 登録日 | 2019-05-16 | | 公開日 | 2020-02-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A suicide enzyme catalyzes multiple reactions for biotin biosynthesis in cyanobacteria.
Nat.Chem.Biol., 16, 2020
|
|
6K38
 
 | |
7X56
 
 | | A CBg-ParM filament with ADP | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ParM/StbA family protein | | 著者 | Koh, A, Ali, S, Robinson, R, Narita, A. | | 登録日 | 2022-03-04 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | A new actin-like filament from Clostridium botulinum exhibits a new system of filament turn over
To Be Published
|
|
7X59
 
 | |
7X55
 
 | |
7X54
 
 | | A CBg-ParM filament with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ParM/StbA family protein | | 著者 | Koh, A, Ali, S, Robinson, R, Narita, A. | | 登録日 | 2022-03-04 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | A new actin-like filament from Clostridium botulinum exhibits a new system of filament turn over
To Be Published
|
|
5YJ9
 
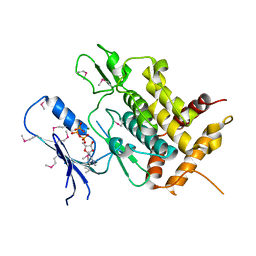 | | Crystal structure of Tribolium castaneum PINK1 kinase domain in complex with AMP-PNP | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase PINK1, ... | | 著者 | Okatsu, K, Sato, Y, Fukai, S. | | 登録日 | 2017-10-09 | | 公開日 | 2018-07-25 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Structural insights into ubiquitin phosphorylation by PINK1.
Sci Rep, 8, 2018
|
|
6JWI
 
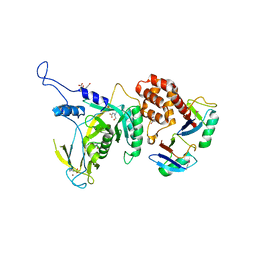 | |
6JWJ
 
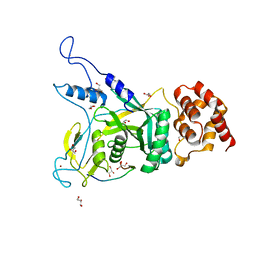 | | Npl4 in complex with Ufd1 | | 分子名称: | GLYCEROL, Nuclear protein localization protein 4, Peptide from Ubiquitin fusion degradation protein 1, ... | | 著者 | Sato, Y, Fukai, S. | | 登録日 | 2019-04-20 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4.
Nat Commun, 10, 2019
|
|
6JWH
 
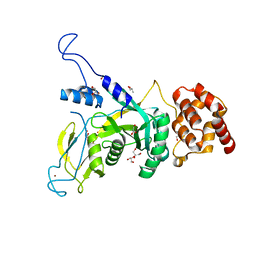 | | Yeast Npl4 zinc finger, MPN and CTD domains | | 分子名称: | GLYCEROL, Nuclear protein localization protein 4, ZINC ION | | 著者 | Sato, Y, Fukai, S. | | 登録日 | 2019-04-20 | | 公開日 | 2019-12-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.72000253 Å) | | 主引用文献 | Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4.
Nat Commun, 10, 2019
|
|
7YNE
 
 | |
3ADE
 
 | |
7Y13
 
 | | Cryo-EM structure of apo-state MrgD-Gi complex (local) | | 分子名称: | PALMITIC ACID, Soluble cytochrome b562,Mas-related G-protein coupled receptor member D | | 著者 | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | 登録日 | 2022-06-06 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
1V79
 
 | | Crystal structures of adenosine deaminase complexed with potent inhibitors | | 分子名称: | 1-{(1R,2S)-1-[2-(2,3,-DICHLOROPHENYL)ETHYL]-2-HYDROXYPROPYL}-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine deaminase, ZINC ION | | 著者 | Kinoshita, T. | | 登録日 | 2003-12-14 | | 公開日 | 2004-12-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-based design and synthesis of non-nucleoside, potent, and orally bioavailable adenosine deaminase inhibitors
J.Med.Chem., 47, 2004
|
|
1WXZ
 
 | | Crystal structure of adenosine deaminase ligated with a potent inhibitor | | 分子名称: | 1-((1R,2S)-1-{2-[2-(4-CHLOROPHENYL)-1,3-BENZOXAZOL-7-YL]ETHYL}-2-HYDROXYPROPYL)-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine deaminase, ZINC ION | | 著者 | Kinoshita, T. | | 登録日 | 2005-02-02 | | 公開日 | 2005-08-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Rational design of non-nucleoside, potent, and orally bioavailable adenosine deaminase inhibitors: predicting enzyme conformational change and metabolism
J.Med.Chem., 48, 2005
|
|
7Y14
 
 | | Cryo-EM structure of MrgD-Gi complex with beta-alanine (local) | | 分子名称: | BETA-ALANINE, PALMITIC ACID, Soluble cytochrome b562,Mas-related G-protein coupled receptor member D | | 著者 | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | 登録日 | 2022-06-06 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7Y12
 
 | | Cryo-EM structure of MrgD-Gi complex with beta-alanine | | 分子名称: | BETA-ALANINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | 登録日 | 2022-06-06 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-02-15 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
7Y15
 
 | | Cryo-EM structure of apo-state MrgD-Gi complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Suzuki, S, Iida, M, Kawamoto, A, Oshima, A. | | 登録日 | 2022-06-06 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Commun Biol, 5, 2022
|
|
1V7A
 
 | |
3A33
 
 | | UbcH5b~Ubiquitin Conjugate | | 分子名称: | GLYCEROL, Ubiquitin, Ubiquitin-conjugating enzyme E2 D2 | | 著者 | Sakata, E, Satoh, T, Yamamoto, S, Yamaguchi, Y, Yagi-Utsumi, M, Kurimoto, E, Wakatsuki, S, Kato, K. | | 登録日 | 2009-06-08 | | 公開日 | 2009-11-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of UbcH5b~Ubiquitin Intermediate: Insight into the Formation of the Self-Assembled E2~Ub Conjugates
Structure, 18, 2010
|
|
2E1W
 
 | |
1V5W
 
 | | Crystal structure of the human Dmc1 protein | | 分子名称: | Meiotic recombination protein DMC1/LIM15 homolog | | 著者 | Kinebuchi, T, Kagawa, W, Enomoto, R, Ikawa, S, Shibata, T, Kurumizaka, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-11-26 | | 公開日 | 2004-05-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein dmc1
Mol.Cell, 14, 2004
|
|
3A2G
 
 | | Crystal Structure of K102C-Myoglobin conjugated with Fluorescein | | 分子名称: | 1-methylpyrrolidine-2,5-dione, GLYCEROL, Myoglobin, ... | | 著者 | Koshiyama, T, Hikage, T, Ueno, T. | | 登録日 | 2009-05-20 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Modification of porous protein crystals in development of biohybrid materials
Bioconjug.Chem., 21, 2010
|
|
