8IM7
 
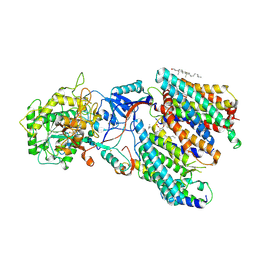 | | Human gamma-secretase treated with ganglioside GM1 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhou, R, Yang, G, Shi, Y. | | 登録日 | 2023-03-06 | | 公開日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Preferential Regulation of Gamma-Secretase-Mediated Cleavage of APP by Ganglioside GM1 Reveals a Potential Therapeutic Target for Alzheimer's Disease.
Adv Sci, 10, 2023
|
|
6EG8
 
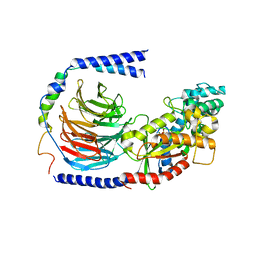 | | Structure of the GDP-bound Gs heterotrimer | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Hilger, D, Liu, X, Aschauer, P, Kobilka, B.K. | | 登録日 | 2018-08-19 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Insights into the Process of GPCR-G Protein Complex Formation.
Cell, 177, 2019
|
|
8EB9
 
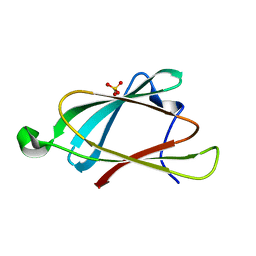 | |
8EBB
 
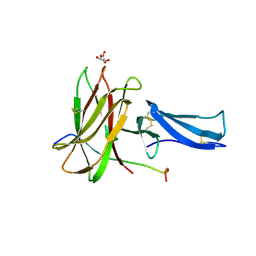 | |
5N0L
 
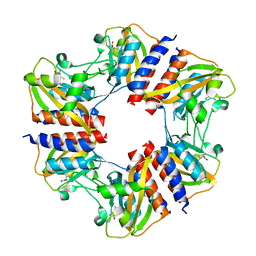 | | The structure of the cofactor binding GAF domain of the nutrient sensor CodY from Clostridium difficile | | 分子名称: | GTP-sensing transcriptional pleiotropic repressor CodY, ISOLEUCINE | | 著者 | Levdikov, V.M, Blagova, E.V, Wilkinson, A.J, Sonenshein, A.L. | | 登録日 | 2017-02-03 | | 公開日 | 2018-02-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Impact of CodY protein on metabolism, sporulation and virulence in Clostridioides difficile ribotype 027.
Plos One, 14, 2019
|
|
7NFX
 
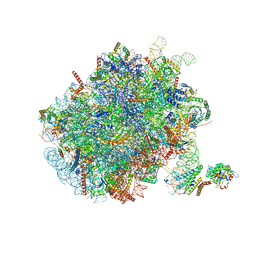 | | Mammalian ribosome nascent chain complex with SRP and SRP receptor in early state A | | 分子名称: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | 著者 | Jomaa, A, Lee, J.H, Shan, S, Ban, N. | | 登録日 | 2021-02-08 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Receptor compaction and GTPase rearrangement drive SRP-mediated cotranslational protein translocation into the ER.
Sci Adv, 7, 2021
|
|
7RCW
 
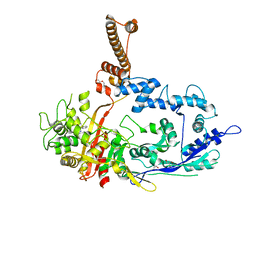 | | Crystal structure of C. difficile penicillin-binding protein 2 in complex with ampicillin | | 分子名称: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2021-07-08 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7RCX
 
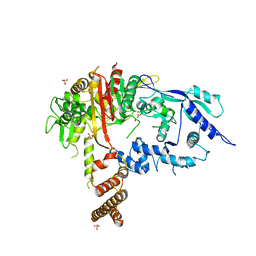 | |
7RCZ
 
 | | Crystal structure of C. difficile SpoVD in complex with ampicillin | | 分子名称: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2021-07-08 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7RCY
 
 | | Crystal structure of C. difficile penicillin-binding protein 2 in complex with ceftobiprole | | 分子名称: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein, ZINC ION | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2021-07-08 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7RD0
 
 | |
4GBY
 
 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-xylose | | 分子名称: | D-xylose-proton symporter, beta-D-xylopyranose, nonyl beta-D-glucopyranoside | | 著者 | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | 登録日 | 2012-07-28 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.808 Å) | | 主引用文献 | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4GBZ
 
 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-glucose | | 分子名称: | D-xylose-proton symporter, beta-D-glucopyranose, nonyl beta-D-glucopyranoside | | 著者 | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | 登録日 | 2012-07-28 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.894 Å) | | 主引用文献 | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4GC0
 
 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to 6-bromo-6-deoxy-D-glucose | | 分子名称: | 6-bromo-6-deoxy-beta-D-glucopyranose, D-xylose-proton symporter, nonyl beta-D-glucopyranoside | | 著者 | Yan, N, Sun, L.F, Zeng, X, Yan, C.Y. | | 登録日 | 2012-07-28 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
6HY0
 
 | | Atomic models of P1, P4 C-terminal fragment and P8 fitted in the bacteriophage phi6 nucleocapsid reconstructed with icosahedral symmetry | | 分子名称: | Major Outer Capsid Protein P8, Major inner protein P1, Packaging Enzyme P4 | | 著者 | El Omari, K, Ilca, S.L, Stuart, D.I, Huiskonen, J.T. | | 登録日 | 2018-10-18 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Multiple liquid crystalline geometries of highly compacted nucleic acid in a dsRNA virus.
Nature, 570, 2019
|
|
5IT3
 
 | | Swirm domain of human Lsd1 | | 分子名称: | Lysine-specific histone demethylase 1A, MAGNESIUM ION | | 著者 | Jeffrey, P.D, Yuan, P. | | 登録日 | 2016-03-16 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A Tlx-interacting peptide of Lsd1 inhibits the proliferation of brain tumor stem cells
To Be Published
|
|
5WY2
 
 | |
6YNO
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_139 | | 分子名称: | 6-[[methyl-[(1-phenylpyrazol-3-yl)methyl]amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | 著者 | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | 登録日 | 2020-04-14 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZD7
 
 | |
6ZCN
 
 | | Crystal structure of YTHDC1 with m6A | | 分子名称: | N6-METHYLADENOSINE-5'-MONOPHOSPHATE, SULFATE ION, YTHDC1 | | 著者 | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | 登録日 | 2020-06-11 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZD4
 
 | |
6ZCM
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_180 | | 分子名称: | 6-[[cyclopropyl-[(7-methoxy-1,3-benzodioxol-5-yl)methyl]amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | 著者 | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | 登録日 | 2020-06-11 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZD3
 
 | | Crystal structure of YTHDC1 M438A mutant | | 分子名称: | DI(HYDROXYETHYL)ETHER, SULFATE ION, YTH domain containing 1 | | 著者 | Bedi, R.K, Li, Y, Caflisch, A. | | 登録日 | 2020-06-13 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZD5
 
 | |
6ZDA
 
 | |
