6S3D
 
 | |
7ZSD
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, local | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, de novo designed binder | | 著者 | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | 登録日 | 2022-05-06 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZSS
 
 | | cryo-EM structure of D614 spike in complex with de novo designed binder | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | 登録日 | 2022-05-08 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZRV
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, full map | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, ... | | 著者 | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | 登録日 | 2022-05-05 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
3UOX
 
 | | Crystal Structure of OTEMO (FAD bound form 2) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, OTEMO | | 著者 | Shi, R, Matte, A, Cygler, M, Lau, P. | | 登録日 | 2011-11-17 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.956 Å) | | 主引用文献 | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UOZ
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 2) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | 著者 | Shi, R, Matte, A, Cygler, M, Lau, P. | | 登録日 | 2011-11-17 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.407 Å) | | 主引用文献 | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UOY
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 1) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO, ... | | 著者 | Shi, R, Matte, A, Cygler, M, Lau, P. | | 登録日 | 2011-11-17 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UOV
 
 | | Crystal Structure of OTEMO (FAD bound form 1) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, OTEMO | | 著者 | Shi, R, Matte, A, Cygler, M, Lau, P. | | 登録日 | 2011-11-17 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.045 Å) | | 主引用文献 | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UP5
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 4) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | 著者 | Shi, R, Matte, A, Cygler, M, Lau, P. | | 登録日 | 2011-11-17 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.453 Å) | | 主引用文献 | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
3UP4
 
 | | Crystal Structure of OTEMO complex with FAD and NADP (form 3) | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OTEMO | | 著者 | Shi, R, Matte, A, Cygler, M, Lau, P. | | 登録日 | 2011-11-17 | | 公開日 | 2012-02-01 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.804 Å) | | 主引用文献 | Cloning, Baeyer-Villiger biooxidations, and structures of the camphor pathway 2-oxo-{Delta}(3)-4,5,5-trimethylcyclopentenylacetyl-coenzyme A monooxygenase of Pseudomonas putida ATCC 17453.
Appl.Environ.Microbiol., 78, 2012
|
|
8BD9
 
 | | Crystal structure of TRIM33 alpha PHD-Bromo domain in complex with 10 | | 分子名称: | 1,3-dimethylbenzimidazol-2-one, CALCIUM ION, E3 ubiquitin-protein ligase TRIM33, ... | | 著者 | Tassone, G, Pozzi, C, Palomba, T. | | 登録日 | 2022-10-18 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Exploiting ELIOT for Scaffold-Repurposing Opportunities: TRIM33 a Possible Novel E3 Ligase to Expand the Toolbox for PROTAC Design.
Int J Mol Sci, 23, 2022
|
|
8BDY
 
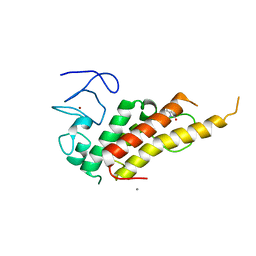 | | Crystal structure of TRIM33 alpha PHD-Bromo domain in complex with 9 | | 分子名称: | 1,3-dimethylbenzimidazol-2-one, CALCIUM ION, E3 ubiquitin-protein ligase TRIM33, ... | | 著者 | Tassone, G, Pozzi, C, Palomba, T. | | 登録日 | 2022-10-20 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Exploiting ELIOT for Scaffold-Repurposing Opportunities: TRIM33 a Possible Novel E3 Ligase to Expand the Toolbox for PROTAC Design.
Int J Mol Sci, 23, 2022
|
|
8BD8
 
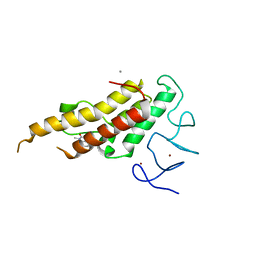 | | Crystal structure of TRIM33 alpha PHD-Bromo domain in complex with 8 | | 分子名称: | 1,3-dimethylbenzimidazol-2-one, CALCIUM ION, E3 ubiquitin-protein ligase TRIM33, ... | | 著者 | Tassone, G, Pozzi, C, Palomba, T. | | 登録日 | 2022-10-18 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Exploiting ELIOT for Scaffold-Repurposing Opportunities: TRIM33 a Possible Novel E3 Ligase to Expand the Toolbox for PROTAC Design.
Int J Mol Sci, 23, 2022
|
|
7XAD
 
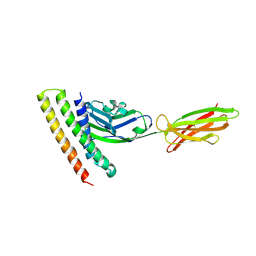 | | Crystal strucutre of PD-L1 and DBL2_02 designed protein binder | | 分子名称: | DBL2_02 binder, Programmed cell death 1 ligand 1 | | 著者 | Liu, K.F, Xu, Z.P, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S.G. | | 登録日 | 2022-03-17 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7XYQ
 
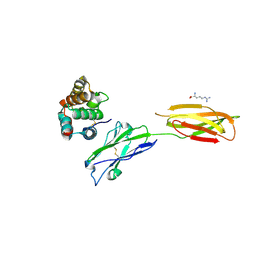 | | Crystal strucutre of PD-L1 and the computationally designed DBL1_03 protein binder | | 分子名称: | ARGININE, CD274 molecule, DBL1_03 | | 著者 | Liu, K, Xu, Z, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S. | | 登録日 | 2022-06-02 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-05-17 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
6VTW
 
 | |
2QX3
 
 | |
2QY1
 
 | | pectate lyase A31G/R236F from Xanthomonas campestris | | 分子名称: | PHOSPHATE ION, Pectate lyase II | | 著者 | Garron, M.L, Shaya, D. | | 登録日 | 2007-08-13 | | 公開日 | 2008-02-26 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Improvement of the thermostability and activity of a pectate lyase by single amino acid substitutions, using a strategy based on melting-temperature-guided sequence alignment.
Appl.Environ.Microbiol., 74, 2008
|
|
2QXZ
 
 | | pectate lyase R236F from Xanthomonas campestris | | 分子名称: | PHOSPHATE ION, pectate lyase II | | 著者 | Garron, M.L, Shaya, D. | | 登録日 | 2007-08-13 | | 公開日 | 2008-03-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Improvement of the thermostability and activity of a pectate lyase by single amino acid substitutions, using a strategy based on melting-temperature-guided sequence alignment.
Appl.Environ.Microbiol., 74, 2008
|
|
1A42
 
 | | HUMAN CARBONIC ANHYDRASE II COMPLEXED WITH BRINZOLAMIDE | | 分子名称: | (4R)-2-(2-ethoxyethyl)-4-(ethylamino)-3,4-dihydro-2H-thieno[3,2-e][1,2]thiazine-6-sulfonamide 1,1-dioxide, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | 著者 | Boriack-Sjodin, P.A, Christianson, D.W. | | 登録日 | 1998-02-10 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structures of murine carbonic anhydrase IV and human carbonic anhydrase II complexed with brinzolamide: molecular basis of isozyme-drug discrimination.
Protein Sci., 7, 1998
|
|
7RWG
 
 | | "Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-43192 | | 分子名称: | (8R)-8-(4-chlorophenyl)-6-(2-methyl-2H-indazol-5-yl)-2-[(2,2,2-trifluoroethyl)amino]-5,8-dihydropyrido[4,3-d]pyrimidin-7(6H)-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Jin, L, Padyana, A.K. | | 登録日 | 2021-08-19 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.97 Å) | | 主引用文献 | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RWH
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-41998 | | 分子名称: | 1,2-ETHANEDIOL, 8-(4-bromophenyl)-6-(4-methoxyphenyl)-2-[2,2,2-tris(fluoranyl)ethylamino]pyrido[4,3-d]pyrimidin-7-ol, CHLORIDE ION, ... | | 著者 | Jin, L, Padyana, A.K. | | 登録日 | 2021-08-19 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RW7
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor Compound 9 | | 分子名称: | (3'R)-2-[(cyclopropylmethyl)amino]-6-(4-methoxyphenyl)-1'-[(1H-pyrazol-5-yl)methyl]-5,6-dihydro-7H-spiro[pyrido[4,3-d]pyrimidine-8,3'-pyrrolidin]-7-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Jin, L, Padyana, A.K. | | 登録日 | 2021-08-19 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.19 Å) | | 主引用文献 | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RW5
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor Compound 1 | | 分子名称: | (3'R)-N-(cyclopropylmethyl)-1'-[(2-fluorophenyl)methyl]-4-methyl-5H,7H-spiro[pyrano[4,3-d]pyrimidine-8,3'-pyrrolidin]-2-amine, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | 著者 | Jin, L, Padyana, A.K. | | 登録日 | 2021-08-19 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7KCF
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-24512 | | 分子名称: | 1,2-ETHANEDIOL, 6-(4-hydroxyphenyl)-5-methyl-2-phenyl-3-(piperidin-1-yl)pyrazolo[1,5-a]pyrimidin-7(4H)-one, GLYCEROL, ... | | 著者 | Padyana, A, Jin, L. | | 登録日 | 2020-10-05 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
