8H2X
 
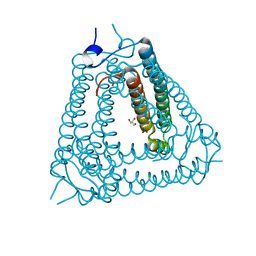 | | Structure of Acb2 | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, p26 | | 著者 | Feng, Y, Cao, X.L. | | 登録日 | 2022-10-07 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
8H39
 
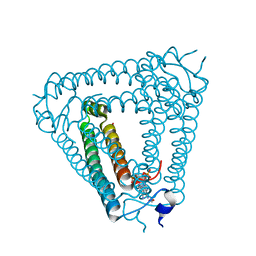 | | Structure of Acb2 complexed with c-di-AMP | | 分子名称: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, 1,2-ETHANEDIOL, p26 | | 著者 | Feng, Y, Cao, X.L. | | 登録日 | 2022-10-08 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
8H2J
 
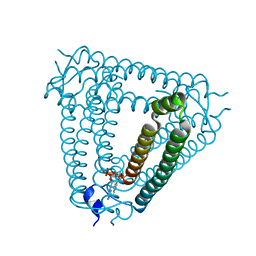 | | Structure of Acb2 complexed with 3',3'-cGAMP | | 分子名称: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, p26 | | 著者 | Feng, Y, Cao, X.L. | | 登録日 | 2022-10-06 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
8GZ4
 
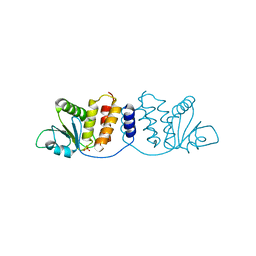 | | Crystal structure of MPXV phosphatase | | 分子名称: | Dual specificity protein phosphatase H1, PHOSPHATE ION | | 著者 | Yang, H.T, Wang, W, Huang, H.J, Ji, X.Y. | | 登録日 | 2022-09-25 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Crystal structure of monkeypox H1 phosphatase, an antiviral drug target.
Protein Cell, 14, 2023
|
|
8HLE
 
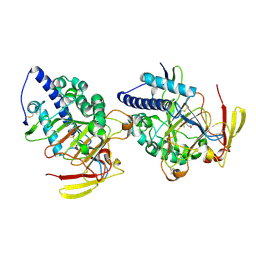 | | Structure of DddY-DMSOP complex | | 分子名称: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, DMSP lyase DddY, ZINC ION | | 著者 | Peng, M, Li, C.Y, Zhang, Y.Z. | | 登録日 | 2022-11-30 | | 公開日 | 2023-10-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
8HLF
 
 | | Crystal structure of DddK-DMSOP complex | | 分子名称: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, MANGANESE (II) ION, Novel protein with potential Cupin domain | | 著者 | Peng, M, Li, C.Y, Zhang, Y.Z. | | 登録日 | 2022-11-30 | | 公開日 | 2023-10-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
7KXT
 
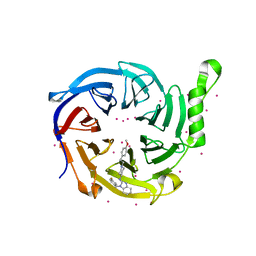 | | Crystal structure of human EED | | 分子名称: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Polycomb protein EED, UNKNOWN ATOM OR ION | | 著者 | Zhu, L, Dong, A, Du, D, Liu, Y, Luo, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2020-12-04 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure-Guided Development of Small-Molecule PRC2 Inhibitors Targeting EZH2-EED Interaction.
J.Med.Chem., 64, 2021
|
|
8WND
 
 | |
6AP7
 
 | |
6AP8
 
 | |
6AP6
 
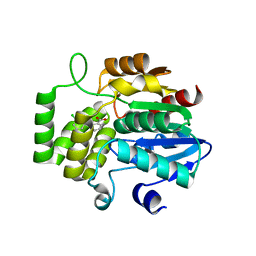 | | Crystal Structure of DAD2 in complex with tolfenamic acid | | 分子名称: | 2-[(3-chloro-2-methylphenyl)amino]benzoic acid, Probable strigolactone esterase DAD2 | | 著者 | Hamiaux, C. | | 登録日 | 2017-08-17 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Inhibition of strigolactone receptors byN-phenylanthranilic acid derivatives: Structural and functional insights.
J. Biol. Chem., 293, 2018
|
|
5EW6
 
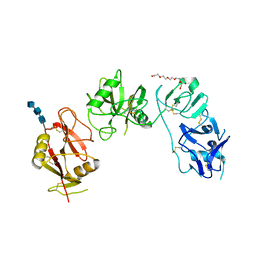 | | Structure of ligand binding region of uPARAP at pH 7.4 without calcium | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type mannose receptor 2, ... | | 著者 | Yuan, C, Huang, M. | | 登録日 | 2015-11-20 | | 公開日 | 2016-08-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Crystal structures of the ligand-binding region of uPARAP: effect of calcium ion binding
Biochem.J., 473, 2016
|
|
5E4K
 
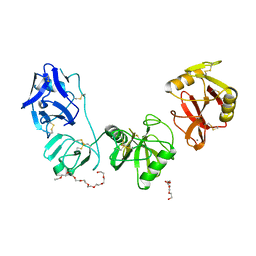 | | Structure of ligand binding region of uPARAP at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, C-type mannose receptor 2, ... | | 著者 | Yuan, C, Huang, M. | | 登録日 | 2015-10-06 | | 公開日 | 2016-08-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Crystal structures of the ligand-binding region of uPARAP: effect of calcium ion binding
Biochem.J., 473, 2016
|
|
5XS3
 
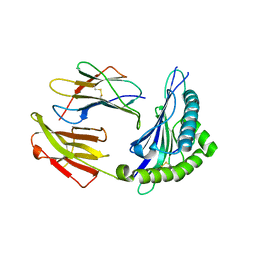 | | Crystal structure of HLA Class I antigen | | 分子名称: | Heavy Chain, Light Chain, P | | 著者 | Wei, P.C, Yang, Y, Liu, Z.X, Luo, Z.Q, Tu, W.Y, Han, J.Y, Deng, Y.H, Yin, L. | | 登録日 | 2017-06-12 | | 公開日 | 2017-09-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Characterization of Autoantigen Presentation by HLA-C*06:02 in Psoriasis
J. Invest. Dermatol., 137, 2017
|
|
4R15
 
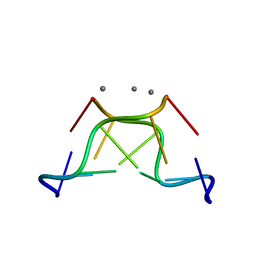 | | High-resolution crystal structure of Z-DNA in complex with Cr3+ cations | | 分子名称: | CHROMIUM ION, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | 著者 | Drozdzal, P, Gilski, M, Kierzek, R, Lomozik, L, Jaskolski, M. | | 登録日 | 2014-08-04 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.97 Å) | | 主引用文献 | High-resolution crystal structure of Z-DNA in complex with Cr(3+) cations.
J.Biol.Inorg.Chem., 20, 2015
|
|
6LI1
 
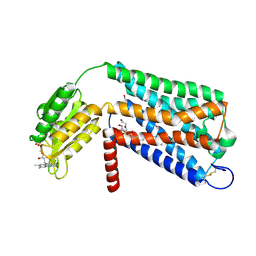 | | Crystal structure of GPR52 ligand free form with flavodoxin fusion | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Flavodoxin, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | 登録日 | 2019-12-10 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LI0
 
 | | Crystal structure of GPR52 in complex with agonist c17 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRATE ANION, Chimera of G-protein coupled receptor 52 and Flavodoxin, ... | | 著者 | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | 登録日 | 2019-12-10 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LI2
 
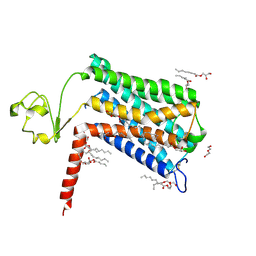 | | Crystal structure of GPR52 ligand free form with rubredoxin fusion | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Rubredoxin, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | 登録日 | 2019-12-10 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
7F5H
 
 | | The crystal structure of RBD-Nanobody complex, DL28 (SC4) | | 分子名称: | GLYCEROL, Nanobody DL28, PHOSPHATE ION, ... | | 著者 | Luo, Z.P, Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | 登録日 | 2021-06-22 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Characterization of a Neutralizing Nanobody With Broad Activity Against SARS-CoV-2 Variants.
Front Microbiol, 13, 2022
|
|
8XVJ
 
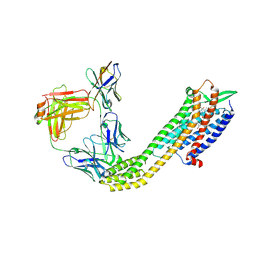 | | Cryo-EM structure of ETAR bound with Macitentan | | 分子名称: | 6-[2-(5-bromanylpyrimidin-2-yl)oxyethoxy]-5-(4-bromophenyl)-~{N}-(propylsulfamoyl)pyrimidin-4-amine, Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, ... | | 著者 | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | 登録日 | 2024-01-15 | | 公開日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVK
 
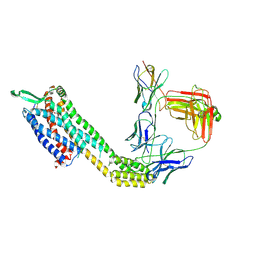 | | Cryo-EM structure of ETAR bound with Ambrisentan | | 分子名称: | (2~{S})-2-(4,6-dimethylpyrimidin-2-yl)oxy-3-methoxy-3,3-diphenyl-propanoic acid, Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, ... | | 著者 | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | 登録日 | 2024-01-15 | | 公開日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.21 Å) | | 主引用文献 | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVL
 
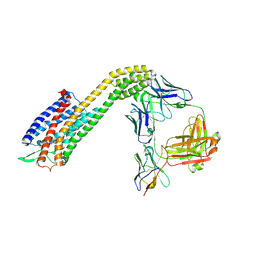 | | Cryo-EM structure of ETAR bound with Zibotentan | | 分子名称: | Endoglucanase H,Endothelin-1 receptor,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, anti-BRIL Fab Light chain, ... | | 著者 | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | 登録日 | 2024-01-15 | | 公開日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVI
 
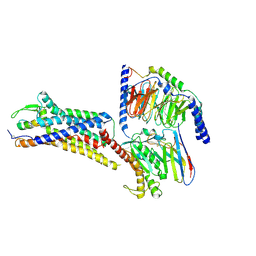 | | Cryo-EM structure of ETAR bound with Endothelin1 | | 分子名称: | Endoglucanase H,Endothelin-1 receptor, Endothelin-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | 登録日 | 2024-01-15 | | 公開日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVH
 
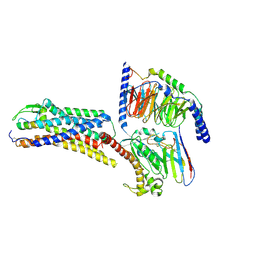 | | Cryo-EM structure of ETBR bound with Endothelin1 | | 分子名称: | Endothelin-1, Exo-alpha-sialidase,Endothelin receptor type B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | 登録日 | 2024-01-15 | | 公開日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVE
 
 | | Cryo-EM structure of ETBR bound with BQ3020 | | 分子名称: | BQ3020, Exo-alpha-sialidase,Endothelin receptor type B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | 登録日 | 2024-01-15 | | 公開日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
