1ZY7
 
 | | Crystal structure of the catalytic domain of an adenosine deaminase that acts on RNA (hADAR2) bound to inositol hexakisphosphate (IHP) | | 分子名称: | INOSITOL HEXAKISPHOSPHATE, RNA-specific adenosine deaminase B1, isoform DRADA2a, ... | | 著者 | Macbeth, M.R, Schubert, H.L, Vandemark, A.P, Lingam, A.T, Hill, C.P, Bass, B.L. | | 登録日 | 2005-06-09 | | 公開日 | 2005-09-13 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing.
Science, 309, 2005
|
|
1Z3Y
 
 | | Structure of Gun4-1 from Thermosynechococcus elongatus | | 分子名称: | putative cytidylyltransferase | | 著者 | Davison, P.A, Schubert, H.L, Reid, J.D, Iorg, C.D, Robinson, H, Hill, C.P, Hunter, C.N. | | 登録日 | 2005-03-14 | | 公開日 | 2005-06-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and Biochemical Characterization of Gun4 Suggests a Mechanism for Its Role in Chlorophyll Biosynthesis(,).
Biochemistry, 44, 2005
|
|
1Z7Q
 
 | | Crystal structure of the 20s proteasome from yeast in complex with the proteasome activator PA26 from Trypanosome brucei at 3.2 angstroms resolution | | 分子名称: | Potential proteasome component C5, Proteasome component C1, Proteasome component C11, ... | | 著者 | Forster, A, Whitby, F.G, Hill, C.P. | | 登録日 | 2005-03-26 | | 公開日 | 2005-08-09 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.22 Å) | | 主引用文献 | The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Mol.Cell, 18, 2005
|
|
4LCB
 
 | | Structure of Vps4 homolog from Acidianus hospitalis | | 分子名称: | CHLORIDE ION, Cell division protein CdvC, Vps4 | | 著者 | Han, H, Hill, C.P, Whitby, F.G, Monroe, N. | | 登録日 | 2013-06-21 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | The Oligomeric State of the Active Vps4 AAA ATPase.
J.Mol.Biol., 426, 2014
|
|
4LGM
 
 | | Crystal Structure of Sulfolobus solfataricus Vps4 | | 分子名称: | CHLORIDE ION, Vps4 AAA ATPase | | 著者 | Han, H, Hill, C.P, Whitby, F.G, Monroe, N. | | 登録日 | 2013-06-28 | | 公開日 | 2013-11-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.711 Å) | | 主引用文献 | The Oligomeric State of the Active Vps4 AAA ATPase.
J.Mol.Biol., 426, 2014
|
|
4R0R
 
 | | Ebolavirus GP Prehairpin Intermediate Mimic | | 分子名称: | eboIZN21 | | 著者 | Clinton, T.R, Weinstock, M.T, Jacobsen, M.T, Szabo-Fresnais, N, Pandya, M.J, Whitby, F.G, Herbert, A.S, Prugar, L.I, McKinnon, R, Hill, C.P, Welch, B.D, Dye, J.M, Eckert, D.M, Kay, M.S. | | 登録日 | 2014-08-01 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Design and characterization of ebolavirus GP prehairpin intermediate mimics as drug targets.
Protein Sci., 24, 2015
|
|
4IOY
 
 | |
4I6M
 
 | | Structure of Arp7-Arp9-Snf2(HSA)-RTT102 subcomplex of SWI/SNF chromatin remodeler. | | 分子名称: | Actin-like protein ARP9, Actin-related protein 7, PHOSPHATE ION, ... | | 著者 | Schubert, H.L, Cairns, B.R, Hill, C.P. | | 登録日 | 2012-11-29 | | 公開日 | 2013-02-13 | | 最終更新日 | 2017-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | Structure of an actin-related subcomplex of the SWI/SNF chromatin remodeler.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4G4S
 
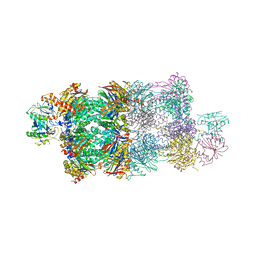 | | Structure of Proteasome-Pba1-Pba2 Complex | | 分子名称: | MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-4-methyl-1-oxopentan-2-yl]-L-leucinamide, Proteasome assembly chaperone 2, ... | | 著者 | Kish-Trier, E, Robinson, H, Stadtmueller, B.M, Hill, C.P. | | 登録日 | 2012-07-16 | | 公開日 | 2012-09-05 | | 最終更新日 | 2013-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Structure of a Proteasome Pba1-Pba2 Complex: IMPLICATIONS FOR PROTEASOME ASSEMBLY, ACTIVATION, AND BIOLOGICAL FUNCTION.
J.Biol.Chem., 287, 2012
|
|
3PSK
 
 | |
3RBQ
 
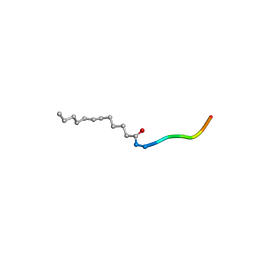 | | Co-crystal structure of human UNC119 (retina gene 4) and an N-terminal Transducin-alpha mimicking peptide | | 分子名称: | Guanine nucleotide-binding protein G(t) subunit alpha-1, Protein unc-119 homolog A | | 著者 | Constantine, R, Whitby, F.G, Hill, C.P, Baehr, W. | | 登録日 | 2011-03-29 | | 公開日 | 2011-06-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | UNC119 is required for G protein trafficking in sensory neurons.
Nat.Neurosci., 14, 2011
|
|
1CMX
 
 | |
2OEX
 
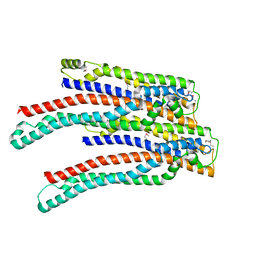 | | Structure of ALIX/AIP1 V Domain | | 分子名称: | Programmed cell death 6-interacting protein | | 著者 | Fisher, R.D, Zhai, Q, Robinson, H, Hill, C.P. | | 登録日 | 2007-01-01 | | 公開日 | 2007-03-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structural and Biochemical Studies of ALIX/AIP1 and Its Role in Retrovirus Budding
Cell(Cambridge,Mass.), 128, 2007
|
|
2OEW
 
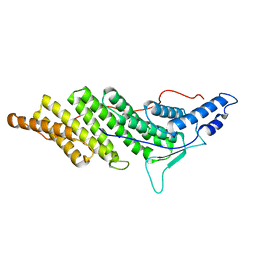 | | Structure of ALIX/AIP1 Bro1 Domain | | 分子名称: | Programmed cell death 6-interacting protein | | 著者 | Fisher, R.D, Zhai, Q, Robinson, H, Hill, C.P. | | 登録日 | 2007-01-01 | | 公開日 | 2007-03-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural and Biochemical Studies of ALIX/AIP1 and Its Role in Retrovirus Budding
Cell(Cambridge,Mass.), 128, 2007
|
|
2R0Y
 
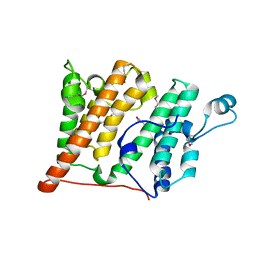 | | Structure of the Rsc4 tandem bromodomain in complex with an acetylated H3 peptide | | 分子名称: | Chromatin structure-remodeling complex protein RSC4, Histone H3 peptide | | 著者 | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | 登録日 | 2007-08-21 | | 公開日 | 2007-10-30 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
2R0V
 
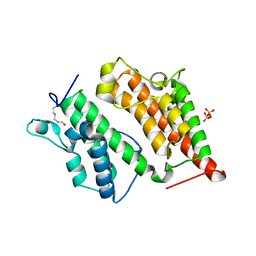 | | Structure of the Rsc4 tandem bromodomain acetylated at K25 | | 分子名称: | Chromatin structure-remodeling complex protein RSC4, SULFATE ION | | 著者 | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | 登録日 | 2007-08-21 | | 公開日 | 2007-10-30 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
2OEV
 
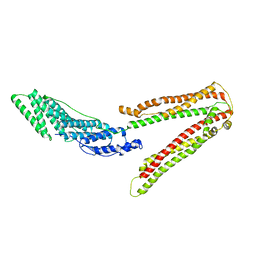 | | Crystal structure of ALIX/AIP1 | | 分子名称: | Programmed cell death 6-interacting protein | | 著者 | Fisher, R.D, Zhai, Q, Robinson, H, Hill, C.P. | | 登録日 | 2007-01-01 | | 公開日 | 2007-03-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural and Biochemical Studies of ALIX/AIP1 and Its Role in Retrovirus Budding
Cell(Cambridge,Mass.), 128, 2007
|
|
2R10
 
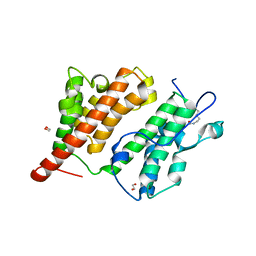 | | Structure of an acetylated Rsc4 tandem bromodomain Histone Chimera | | 分子名称: | 1,2-ETHANEDIOL, Chromatin structure-remodeling complex protein RSC4, LINKER, ... | | 著者 | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | 登録日 | 2007-08-21 | | 公開日 | 2007-10-30 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
2R0S
 
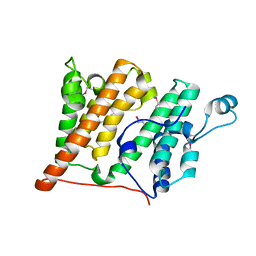 | | Crystal Structure of the Rsc4 tandem bromodomain | | 分子名称: | Chromatin structure-remodeling complex protein RSC4 | | 著者 | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | 登録日 | 2007-08-21 | | 公開日 | 2007-10-30 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
2R3C
 
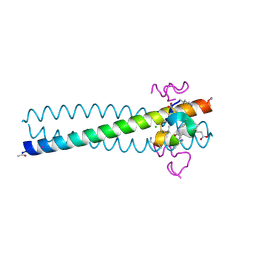 | | Structure of the gp41 N-peptide in complex with the HIV entry inhibitor PIE1 | | 分子名称: | CHLORIDE ION, HIV entry inhibitor PIE1, YTTRIUM (III) ION, ... | | 著者 | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | 登録日 | 2007-08-29 | | 公開日 | 2007-10-02 | | 最終更新日 | 2018-08-08 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R5B
 
 | | Structure of the gp41 N-trimer in complex with the HIV entry inhibitor PIE7 | | 分子名称: | HIV entry inhibitor PIE7, SULFATE ION, gp41 N-peptide | | 著者 | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | 登録日 | 2007-09-03 | | 公開日 | 2007-10-02 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R5D
 
 | | Structure of the gp41 N-trimer in complex with the HIV entry inhibitor PIE7 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, HIV entry inhibitor PIE7, ... | | 著者 | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | 登録日 | 2007-09-03 | | 公開日 | 2007-10-02 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1M9Y
 
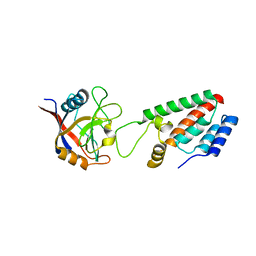 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) M-type H87A,G89A Complex. | | 分子名称: | Cyclophilin A, HIV-1 Capsid | | 著者 | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | 登録日 | 2002-07-30 | | 公開日 | 2003-05-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
1O06
 
 | | Crystal structure of the Vps27p Ubiquitin Interacting Motif (UIM) | | 分子名称: | Vacuolar protein sorting-associated protein VPS27, ZINC ION | | 著者 | Fisher, R.D, Wang, B, Alam, S.L, Higginson, D.S, Rich, R, Myszka, D, Sundquist, W.I, Hill, C.P. | | 登録日 | 2003-02-20 | | 公開日 | 2003-07-22 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure and ubiquitin binding of the ubiquitin-interacting motif.
J.Biol.Chem., 278, 2003
|
|
1M9E
 
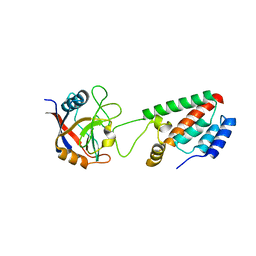 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) M-type H87A Complex. | | 分子名称: | Cyclophilin A, HIV-1 Capsid | | 著者 | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | 登録日 | 2002-07-28 | | 公開日 | 2003-05-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
