1JNP
 
 | | Crystal Structure of Murine Tcl1 at 2.5 Resolution | | 分子名称: | T-CELL LEUKEMIA/LYMPHOMA PROTEIN 1A | | 著者 | Petock, J.M, Torshin, I.Y, Wang, Y.F, DuBois, G.C, Croce, C.M, Harrison, R.W, Weber, I.T. | | 登録日 | 2001-07-24 | | 公開日 | 2001-11-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of murine Tcl1 at 2.5 A resolution and implications for the TCL oncogene family.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1K1T
 
 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | 分子名称: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN, SULFATE ION | | 著者 | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2001-09-25 | | 公開日 | 2002-07-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
1K2C
 
 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | 分子名称: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | 著者 | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, W Harrison, R, Weber, I.T. | | 登録日 | 2001-09-26 | | 公開日 | 2002-07-10 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
1K1U
 
 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | 分子名称: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | 著者 | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, W Harrison, R, Weber, I.T. | | 登録日 | 2001-09-25 | | 公開日 | 2002-07-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
1K2B
 
 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | 分子名称: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | 著者 | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, W Harrison, R, Weber, I.T. | | 登録日 | 2001-09-26 | | 公開日 | 2002-07-10 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
3LJP
 
 | | Crystal structure of choline oxidase V464A mutant | | 分子名称: | Choline oxidase, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | 著者 | Finnegan, S, Agniswamy, J, Weber, I.T, Giovanni, G. | | 登録日 | 2010-01-26 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Role of valine 464 in the flavin oxidation reaction catalyzed by choline oxidase.
Biochemistry, 49, 2010
|
|
3NYC
 
 | |
3NU6
 
 | |
3NU5
 
 | | Crystal Structure of HIV-1 Protease Mutant I50V with Antiviral Drug Amprenavir | | 分子名称: | ACETATE ION, CHLORIDE ION, SODIUM ION, ... | | 著者 | Wang, Y.-F, Shen, C.H, Weber, I.T. | | 登録日 | 2010-07-06 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
3NYF
 
 | |
3NUJ
 
 | |
3NYE
 
 | |
3NU4
 
 | | Crystal Structure of HIV-1 Protease Mutant V32I with Antiviral Drug Amprenavir | | 分子名称: | CHLORIDE ION, SODIUM ION, protease, ... | | 著者 | Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | 登録日 | 2010-07-06 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
3NUO
 
 | |
3NU9
 
 | |
3NU3
 
 | | Wild Type HIV-1 Protease with Antiviral Drug Amprenavir | | 分子名称: | CHLORIDE ION, GLYCEROL, Protease, ... | | 著者 | Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | 登録日 | 2010-07-06 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
3NNE
 
 | | Crystal structure of choline oxidase S101A mutant | | 分子名称: | ACETATE ION, Choline oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Wang, Y.-F, Finnegan, S, Yuan, H, Orville, A.M, Weber, I.T, Gadda, G. | | 登録日 | 2010-06-23 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Structural and kinetic studies on the Ser101Ala variant of choline oxidase: Catalysis by compromise.
Arch.Biochem.Biophys., 501, 2010
|
|
3MWS
 
 | | Crystal Structure of Group N HIV-1 Protease | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, HIV-1 Protease | | 著者 | Sayer, J.M, Agniswamy, J, Weber, I.T, Louis, J.M. | | 登録日 | 2010-05-06 | | 公開日 | 2011-03-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Autocatalytic maturation, physical/chemical properties, and crystal structure of group N HIV-1 protease: relevance to drug resistance.
Protein Sci., 19, 2010
|
|
3OK9
 
 | | Crystal structure of wild-type HIV-1 protease with new oxatricyclic designed inhibitor GRL-0519A | | 分子名称: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2010-08-24 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Probing Multidrug-Resistance and Protein-Ligand Interactions with Oxatricyclic Designed Ligands in HIV-1 Protease Inhibitors.
Chemmedchem, 5, 2010
|
|
3GJR
 
 | | Caspase-3 Binds Diverse P4 Residues in Peptides | | 分子名称: | Caspase-3 subunit p12, Caspase-3 subunit p17, GLYCEROL, ... | | 著者 | Fang, B, Fu, G, Agniswamy, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2009-03-09 | | 公開日 | 2009-03-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling.
Apoptosis, 14, 2009
|
|
3GJT
 
 | | Caspase-3 Binds Diverse P4 Residues in Peptides | | 分子名称: | Caspase-3 subunit p12, Caspase-3 subunit p17, peptide inhibitor | | 著者 | Fang, B, Fu, G, Agniswamy, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2009-03-09 | | 公開日 | 2009-03-24 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling.
Apoptosis, 14, 2009
|
|
3EC0
 
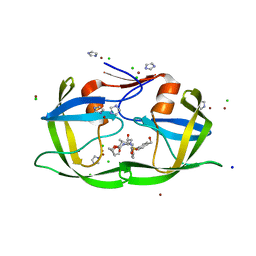 | | High Resolution HIV-2 Protease Structure in Complex with Antiviral Inhibitor GRL-06579A | | 分子名称: | (3AS,5R,6AR)-HEXAHYDRO-2H-CYCLOPENTA[B]FURAN-5-YL (2S,3S)-3-HYDROXY-4-(4-(HYDROXYMETHYL)-N-ISOBUTYLPHENYLSULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, CHLORIDE ION, IMIDAZOLE, ... | | 著者 | Kovalevsky, A.Y, Weber, I.T. | | 登録日 | 2008-08-28 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Structural evidence for effectiveness of darunavir and two related antiviral inhibitors against HIV-2 protease
J.Mol.Biol., 384, 2008
|
|
3GJS
 
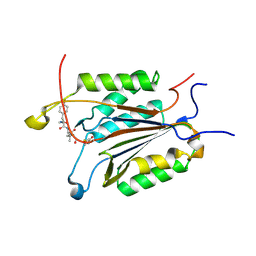 | | Caspase-3 Binds Diverse P4 Residues in Peptides | | 分子名称: | Ac-YVAD-Cho inhibitor, Caspase-3 subunit p12, Caspase-3 subunit p17 | | 著者 | Fang, B, Fu, G, Agniswamy, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2009-03-09 | | 公開日 | 2009-03-24 | | 最終更新日 | 2011-08-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling.
Apoptosis, 14, 2009
|
|
3EBZ
 
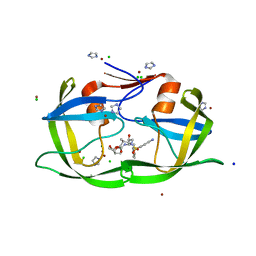 | | High Resolution HIV-2 Protease Structure in Complex with Clinical Drug Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, IMIDAZOLE, ... | | 著者 | Kovalevsky, A.Y, Weber, I.T. | | 登録日 | 2008-08-28 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural evidence for effectiveness of darunavir and two related antiviral inhibitors against HIV-2 protease
J.Mol.Biol., 384, 2008
|
|
3ECG
 
 | | High Resolution HIV-2 Protease Structure in Complex with Antiviral Inhibitor GRL-98065 | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, CHLORIDE ION, IMIDAZOLE, ... | | 著者 | Kovalevsky, A.Y, Weber, I.T. | | 登録日 | 2008-08-29 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Structural evidence for effectiveness of darunavir and two related antiviral inhibitors against HIV-2 protease
J.Mol.Biol., 384, 2008
|
|
