4FA5
 
 | |
6L42
 
 | |
8HNM
 
 | | CXCR3-DNGi complex activated by VUF11222 | | 分子名称: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Jiao, H.Z, Hu, H.L. | | 登録日 | 2022-12-08 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNN
 
 | | Structure of CXCR3 complexed with antagonist SCH546738 | | 分子名称: | 3-azanyl-6-chloranyl-5-[(3S)-4-[1-[(4-chlorophenyl)methyl]piperidin-4-yl]-3-ethyl-piperazin-1-yl]pyrazine-2-carboxamide, CHOLESTEROL, Nb6, ... | | 著者 | Jiao, H.Z, Hu, H.L. | | 登録日 | 2022-12-08 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNK
 
 | | CXCR3-DNGi complex activated by CXCL11 | | 分子名称: | C-X-C motif chemokine 11, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Jiao, H.Z, Hu, H.L. | | 登録日 | 2022-12-08 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNL
 
 | | CXCR3-DNGi complex activated by PS372424 | | 分子名称: | (3S)-N-[(2S)-5-carbamimidamido-1-(cyclohexylmethylamino)-1-oxidanylidene-pentan-2-yl]-2-(4-oxidanylidene-4-phenyl-butanoyl)-3,4-dihydro-1H-isoquinoline-3-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Jiao, H.Z, Hu, H.L. | | 登録日 | 2022-12-08 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8X2J
 
 | |
5OVQ
 
 | |
1Z1Z
 
 | |
6MAE
 
 | |
1TMB
 
 | | MOLECULAR BASIS FOR THE INHIBITION OF HUMAN ALPHA-THROMBIN BY THE MACROCYCLIC PEPTIDE CYCLOTHEONAMIDE A | | 分子名称: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRUGEN, ... | | 著者 | Qiu, X, Padmanabhan, K.P, Maryanoff, B.E, Tulinsky, A. | | 登録日 | 1993-05-27 | | 公開日 | 1994-01-31 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular basis for the inhibition of human alpha-thrombin by the macrocyclic peptide cyclotheonamide A.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
4FAN
 
 | |
6PEU
 
 | |
6PE3
 
 | |
8GTH
 
 | |
7XCT
 
 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 2:1 complex | | 分子名称: | DNA (145-MER), Histone H2A, Histone H2B type 1-K, ... | | 著者 | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | 登録日 | 2022-03-25 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XD1
 
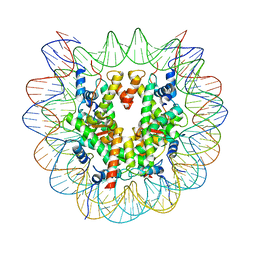 | | cryo-EM structure of unmodified nucleosome | | 分子名称: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | 著者 | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | 登録日 | 2022-03-26 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XCR
 
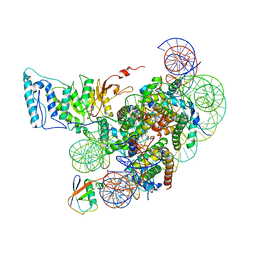 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 1:1 complex | | 分子名称: | DNA (146-MER), Histone H2A, Histone H2B type 1-K, ... | | 著者 | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | 登録日 | 2022-03-25 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XD0
 
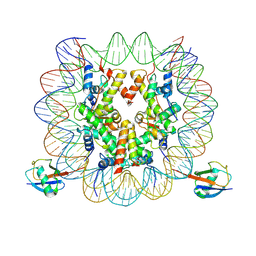 | | cryo-EM structure of H2BK34ub nucleosome | | 分子名称: | DNA (146-MER), Histone H2A, Histone H2B type 1-K, ... | | 著者 | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | 登録日 | 2022-03-26 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
3IE9
 
 | |
3IEA
 
 | |
8SPP
 
 | |
7JIF
 
 | | HRAS A59T GppNHp | | 分子名称: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | 著者 | Johnson, C.W, Haigis, K.M. | | 登録日 | 2020-07-23 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.757 Å) | | 主引用文献 | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
7JII
 
 | | HRAS A59E GDP | | 分子名称: | CALCIUM ION, GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Johnson, C.W, Haigis, K.M. | | 登録日 | 2020-07-23 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.532 Å) | | 主引用文献 | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
7JIH
 
 | | HRAS A59E GppNHp | | 分子名称: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | 著者 | Johnson, C.W, Haigis, K.M. | | 登録日 | 2020-07-23 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.989 Å) | | 主引用文献 | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
