5YQB
 
 | | Crystal structure of E.coli aminopeptidase N in complex with Puromycin | | 分子名称: | (2R,3R,4S,5S)-4-AMINO-2-[6-(DIMETHYLAMINO)-9H-PURIN-9-YL]-5-(HYDROXYMETHYL)TETRAHYDRO-3-FURANOL, Aminopeptidase N, GLYCEROL, ... | | 著者 | Marapaka, A.K, Ganji, R.J, Reddi, R, Addlagatta, A. | | 登録日 | 2017-11-06 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Puromycin, a selective inhibitor of PSA acts as a substrate for other M1 family aminopeptidases: Biochemical and structural basis
Int.J.Biol.Macromol., 165, 2020
|
|
5YR5
 
 | | Human methionine aminopeptidase type 1b (F309L mutant) in complex with Ovalicin | | 分子名称: | 3,4-DIHYDROXY-2-METHOXY-4-METHYL-3-[2-METHYL-3-(3-METHYL-BUT-2-ENYL) -OXIRANYL]-CYCLOHEXANONE, COBALT (II) ION, Methionine aminopeptidase 1, ... | | 著者 | Pillalamarri, V, Arya, T, Addlagatta, A. | | 登録日 | 2017-11-08 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of natural product ovalicin sensitive type 1 methionine aminopeptidases: molecular and structural basis.
Biochem. J., 476, 2019
|
|
5Y5I
 
 | | Time-resolved SFX structure of cytochrome P450nor: 20 ms after photo-irradiation of caged NO in the presence of NADH (NO-bound state), light data | | 分子名称: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | 著者 | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5X
 
 | | V/A-type ATPase/synthase from Thermus thermophilus, rotational state 1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | 著者 | Nakanishi, A, Kishikawa, J, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | 登録日 | 2017-08-10 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Cryo EM structure of intact rotary H+-ATPase/synthase from Thermus thermophilus
Nat Commun, 9, 2018
|
|
5XZH
 
 | | Vitamin D receptor with a synthetic ligand ADRO2 | | 分子名称: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,6R)-6-(1-adamantyl)-6-oxidanyl-hex-4-yn-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | 著者 | Otero, R, Numoto, N, Ikura, T, Yamada, S, Mourino, A, Makishima, M, Ito, N. | | 登録日 | 2017-07-12 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 25 S-Adamantyl-23-yne-26,27-dinor-1 alpha ,25-dihydroxyvitamin D3: Synthesis, Tissue Selective Biological Activities, and X-ray Crystal Structural Analysis of Its Vitamin D Receptor Complex.
J. Med. Chem., 61, 2018
|
|
6T07
 
 | | Crystal structure of YTHDC1 with fragment 20 (DHU_DC1_134) | | 分子名称: | SULFATE ION, YTH domain-containing protein 1, ~{N}-[(2~{S})-pyrrolidin-2-yl]-1~{H}-1,2,4-triazol-5-amine | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-10-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
4C2E
 
 | | Crystal structure of the protease CtpB(S309A) present in a resting state | | 分子名称: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB | | 著者 | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | 登録日 | 2013-08-17 | | 公開日 | 2013-12-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
6T0X
 
 | | Crystal structure of YTHDC1 with fragment 22 (ACA_DC1_001) | | 分子名称: | (3~{S})-~{N}-methylpyrrolidine-3-carboxamide, SULFATE ION, YTHDC1 | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-10-03 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T11
 
 | | Crystal structure of YTHDC1 with fragment 29 (DHU_DC1_218) | | 分子名称: | N-methyl-1H-indole-7-carboxamide, SULFATE ION, YTHDC1 | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-10-03 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
4C59
 
 | | Structure of GAK kinase in complex with nanobody (NbGAK_4) | | 分子名称: | (2Z,3E)-2,3'-BIINDOLE-2',3(1H,1'H)-DIONE 3-{O-[(3R)-3,4-DIHYDROXYBUTYL]OXIME}, Cyclin-G-associated kinase, NANOBODY | | 著者 | Chaikuad, A, Keates, T, Allerston, C.K, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Muller-Knapp, S. | | 登録日 | 2013-09-10 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of cyclin G-associated kinase (GAK) trapped in different conformations using nanobodies.
Biochem. J., 459, 2014
|
|
5Y1Q
 
 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(3-chlorobenzyl)ureido)-N-hydroxy-4-methylpentanamide | | 分子名称: | (2S)-2-[(3-chlorophenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, M1 family aminopeptidase, MAGNESIUM ION, ... | | 著者 | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | 登録日 | 2017-07-21 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Development of peptidomimetic hydroxamates as PfA-M1 and PfA-M17 dual inhibitors: Biological evaluation and structural characterization by cocrystallization
Chin.Chem.Lett., 33, 2022
|
|
5Y25
 
 | | EGFR kinase domain mutant (T790M/L858R) with covalent ligand NS-062 | | 分子名称: | (2R)-N-[4-[(3-chloranyl-4-fluoranyl-phenyl)amino]-7-(3-morpholin-4-ylpropoxy)quinazolin-6-yl]-1-(2-fluoranylethanoyl)pyrrolidine-2-carboxamide, Epidermal growth factor receptor | | 著者 | Shiroishi, M, Abe, Y, Caaveiro, J.M.M, Sakamoto, S, Morimoto, S, Fuchida, H, Shindo, N, Ojida, A. | | 登録日 | 2017-07-24 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.102 Å) | | 主引用文献 | Selective and reversible modification of kinase cysteines with chlorofluoroacetamides.
Nat.Chem.Biol., 15, 2019
|
|
4C2D
 
 | | Crystal structure of the protease CtpB in an active state | | 分子名称: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB, PEPTIDE1, PEPTIDE2 | | 著者 | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | 登録日 | 2013-08-17 | | 公開日 | 2013-12-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
1RGQ
 
 | | M9A HCV Protease complex with pentapeptide keto-amide inhibitor | | 分子名称: | N-(PYRAZIN-2-YLCARBONYL)LEUCYLISOLEUCYL-N~1~-{1-[2-({1-CARBOXY-2-[4-(PHOSPHONOOXY)PHENYL]ETHYL}AMINO)-1,1-DIHYDROXY-2-OXOETHYL]BUT-3-ENYL}-3-CYCLOHEXYLALANINAMIDE, NS3 Protease, NS4A peptide, ... | | 著者 | Liu, Y, Stoll, V.S, Richardson, P.L, Saldivar, A, Klaus, J.L, Molla, A, Kohlbrenner, W, Kati, W.M. | | 登録日 | 2003-11-12 | | 公開日 | 2004-10-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Hepatitis C NS3 protease inhibition by peptidyl-alpha-ketoamide inhibitors: kinetic mechanism and structure.
Arch.Biochem.Biophys., 421, 2004
|
|
5YH4
 
 | |
6SRV
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021144 | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | 登録日 | 2019-09-06 | | 公開日 | 2020-06-10 | | 最終更新日 | 2020-09-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
5YP4
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with Lys-Pro from Pseudoxanthomonas mexicana WO24 | | 分子名称: | Dipeptidyl aminopeptidase 4, GLYCEROL, LYSINE, ... | | 著者 | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | 登録日 | 2017-11-01 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
4BNG
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-pentyl- 2-phenoxyphenol | | 分子名称: | 5-PENTYL-2-PHENOXYPHENOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | 著者 | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | 登録日 | 2013-05-15 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
3W30
 
 | | Structual basis for the recognition of Ubc13 by the Shigella flexneri effector OspI | | 分子名称: | ORF169b | | 著者 | Nishide, A, Kim, M, Takagi, K, Sasakawa, C, Mizushima, T. | | 登録日 | 2012-12-07 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Structural Basis for the Recognition of Ubc13 by the Shigella flexneri Effector OspI.
J.Mol.Biol., 425, 2013
|
|
4XWH
 
 | | Crystal structure of the human N-acetyl-alpha-glucosaminidase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-N-acetylglucosaminidase, ... | | 著者 | Birrane, G, Meiyappan, M, Dassier, A. | | 登録日 | 2015-01-28 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structural characterization of the alpha-N-acetylglucosaminidase, a key enzyme in the pathogenesis of Sanfilippo syndrome B.
J.Struct.Biol., 205, 2019
|
|
5XXM
 
 | | Crystal structure of GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with gluconolactone | | 分子名称: | D-glucono-1,5-lactone, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | 著者 | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | 登録日 | 2017-07-04 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
5YQW
 
 | | Structure and function of a novel periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria | | 分子名称: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NICKEL (II) ION, ... | | 著者 | Suginta, W, Sritho, N, Ranok, A, Kitaoku, Y, Bulmer, D.M, van den Berg, B, Fukamizo, T. | | 登録日 | 2017-11-08 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Structure and function of a novel periplasmic chitooligosaccharide-binding protein from marineVibriobacteria.
J. Biol. Chem., 293, 2018
|
|
6SYZ
 
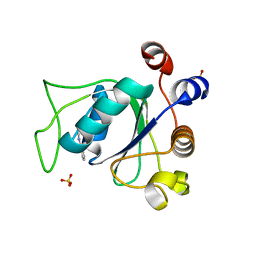 | | Crystal structure of YTHDC1 with fragment 1 (DHU_DC1_141) | | 分子名称: | SULFATE ION, YTH domain-containing protein 1, ~{N}-methylthieno[3,2-d]pyrimidin-4-amine | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-10-01 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
5XZE
 
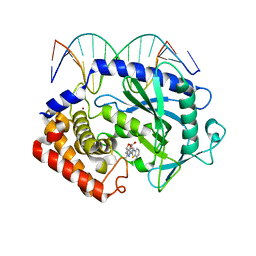 | | Mouse cGAS bound to the inhibitor RU332 | | 分子名称: | (3R)-3-[1-(3H-1lambda~4~,3-benzothiazol-2-yl)-5-hydroxy-3-methyl-1H-pyrazol-4-yl]-2-benzofuran-1(3H)-one, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*G)-3'), ... | | 著者 | Vincent, J, Adura, C, Gao, P, Luz, A, Lama, L, Asano, Y, Okamoto, R, Imaeda, T, Aida, J, Rothamel, K, Gogakos, T, Steinberg, J, Reasoner, S, Aso, K, Tuschl, T, Patel, D.J, Glickman, J.F, Ascano, M. | | 登録日 | 2017-07-12 | | 公開日 | 2017-10-11 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.177 Å) | | 主引用文献 | Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Nat Commun, 8, 2017
|
|
5XZT
 
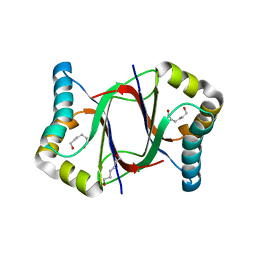 | | C-terminal peptide depleted mutant of hydroxynitrile lyase from Passiflora edulis (PeHNL) | | 分子名称: | HEXANE-1,6-DIOL, Hydroxynitrile lyase | | 著者 | Motojima, F, Nuylert, A, Asano, Y. | | 登録日 | 2017-07-13 | | 公開日 | 2017-11-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structure and catalytic mechanism of hydroxynitrile lyase from passion fruit, Passiflora edulis
FEBS J., 285, 2018
|
|
