4FN8
 
 | | Crystal structure of the Mtb enoyl CoA isomerase (Rv0632c)in complex with acetoacetyl CoA | | 分子名称: | ACETOACETYL-COENZYME A, Enoyl-CoA hydratase/isomerase family protein, SULFATE ION | | 著者 | Bruning, J.B, Gao, N, Hernandez, E.D, Li, H, Dang, N, Hung, L.W, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2012-06-19 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.831 Å) | | 主引用文献 | Crystal structure and mechanism of the prokaryotic enoyl CoA isomerase (ECI)
To be Published
|
|
4FNB
 
 | | Crystal structure of the Mtb enoyl CoA isomerase (Rv0632c) in complex with hydroxybutyrl CoA | | 分子名称: | 3-HYDROXYBUTANOYL-COENZYME A, Enoyl-CoA hydratase/isomerase family protein, SULFATE ION | | 著者 | Bruning, J.B, Gao, N, Hernandez, E.D, Li, H, Dang, N, Hung, L.W, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2012-06-19 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure and Mechanism of the Prokaryotic Enoyl CoA Isomerase
To be Published
|
|
3Q6G
 
 | | Crystal structure of Fab of rhesus mAb 2.5B specific for quaternary neutralizing epitope of HIV-1 gp120 | | 分子名称: | Heavy chain of Fab of rhesus mAb 2.5B, Light chain of Fab of rhesus mAb 2.5B | | 著者 | Spurrier, B, Sampson, J, Totrov, M, Li, H, O'Neal, T, William, C, Robinson, J, Gorny, M.K, Zolla-Pazner, S, Kong, X.P. | | 登録日 | 2010-12-31 | | 公開日 | 2011-05-25 | | 最終更新日 | 2013-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | Structural Analysis of Human and Macaque mAbs 2909 and 2.5B: Implications for the Configuration of the Quaternary Neutralizing Epitope of HIV-1 gp120.
Structure, 19, 2011
|
|
2HVC
 
 | | The Crystal Structure of Ligand-binding Domain (LBD) of human Androgen Receptor in Complex with a selective modulator LGD2226 | | 分子名称: | 6-[BIS(2,2,2-TRIFLUOROETHYL)AMINO]-4-(TRIFLUOROMETHYL)QUINOLIN-2(1H)-ONE, Androgen receptor | | 著者 | Wang, F, Liu, X.-Q, Li, H, Liang, K.-N, Miner, J.N, Hong, M, Kallel, E.A, van Oeveren, A, Zhi, L, Jiang, T. | | 登録日 | 2006-07-28 | | 公開日 | 2007-07-31 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the ligand-binding domain (LBD) of human androgen receptor in complex with a selective modulator LGD2226
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
2I0M
 
 | | Crystal structure of the phosphate transport system regulatory protein PhoU from Streptococcus pneumoniae | | 分子名称: | Phosphate transport system protein phoU, ZINC ION | | 著者 | Zhang, R, Li, H, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-08-10 | | 公開日 | 2006-09-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the phosphate transport system regulatory protein PhoU from Streptococcus pneumoniae
To be Published, 2006
|
|
6L5Z
 
 | |
3QOK
 
 | |
3OOP
 
 | | The structure of a protein with unknown function from Listeria innocua Clip11262 | | 分子名称: | Lin2960 protein | | 著者 | Fan, Y, Li, H, Zhou, Y, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-08-31 | | 公開日 | 2010-09-22 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | The structure of a protein with unknown function from Listeria innocua Clip11262
To be Published
|
|
4EJO
 
 | | Crystal structure of padr family transcriptional regulator from Eggerthella lenta DSM 2243 | | 分子名称: | ETHANOL, Transcriptional regulator, PadR-like family | | 著者 | Chang, C, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-04-06 | | 公開日 | 2012-04-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of padr family transcriptional regulator from Eggerthella lenta DSM 2243
TO BE PUBLISHED
|
|
4FND
 
 | | Crystal structure of the Mtb enoyl CoA isomerase in complex with hydroxyhexanoyl CoA | | 分子名称: | (S)-3-Hydroxyhexanoyl-CoA, Enoyl-CoA hydratase/isomerase family protein, SULFATE ION | | 著者 | Bruning, J.B, Gao, N, Hernandez, E.D, Li, H, Dang, N, Hung, L.W, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2012-06-19 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure and mechanism of the prokaryotic enoyl CoA isomerase
To be Published
|
|
3Q6F
 
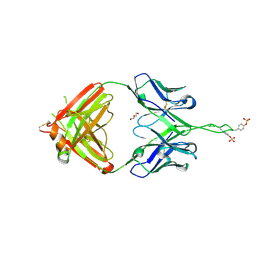 | | Crystal structure of Fab of human mAb 2909 specific for quaternary neutralizing epitope of HIV-1 gp120 | | 分子名称: | DI(HYDROXYETHYL)ETHER, Heavy chain of Fab of human mAb 2909, Light chain of Fab of human mAb 2909 | | 著者 | Spurrier, B, Sampson, J, Totrov, M, Li, H, O'Neal, T, William, C, Robinson, J, Gorny, M.K, Zolla-Pazner, S, Kong, X.P. | | 登録日 | 2010-12-31 | | 公開日 | 2011-05-25 | | 最終更新日 | 2011-10-12 | | 実験手法 | X-RAY DIFFRACTION (3.192 Å) | | 主引用文献 | Structural Analysis of Human and Macaque mAbs 2909 and 2.5B: Implications for the Configuration of the Quaternary Neutralizing Epitope of HIV-1 gp120.
Structure, 19, 2011
|
|
4FYZ
 
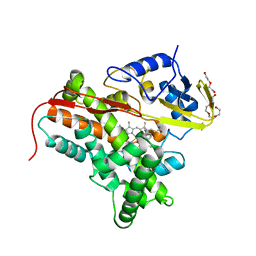 | | Crystal Structure of Nitrosyl Cytochrome P450cin | | 分子名称: | 1,3,3-TRIMETHYL-2-OXABICYCLO[2.2.2]OCTANE, DI(HYDROXYETHYL)ETHER, NITRIC OXIDE, ... | | 著者 | Madrona, Y, Tripathi, S.M, Li, H, Poulos, T.L. | | 登録日 | 2012-07-05 | | 公開日 | 2012-07-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Crystal structures of substrate-free and nitrosyl cytochrome p450cin: implications for o(2) activation.
Biochemistry, 51, 2012
|
|
4G3R
 
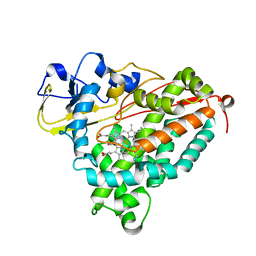 | | Crystal Structure of Nitrosyl Cytochrome P450cam | | 分子名称: | CAMPHOR, Camphor 5-monooxygenase, NITRIC OXIDE, ... | | 著者 | Madrona, Y, Tripathi, S.M, Li, H, Poulos, T.L. | | 登録日 | 2012-07-15 | | 公開日 | 2012-08-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of substrate-free and nitrosyl cytochrome p450cin: implications for o(2) activation.
Biochemistry, 51, 2012
|
|
2YSF
 
 | | Solution structure of the fourth WW domain from the human E3 ubiquitin-protein ligase Itchy homolog, ITCH | | 分子名称: | E3 ubiquitin-protein ligase Itchy homolog | | 著者 | Ohnishi, S, Li, H, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-03 | | 公開日 | 2007-10-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the fourth WW domain from the human E3 ubiquitin-protein ligase Itchy homolog, ITCH
To be Published
|
|
3B79
 
 | |
3B48
 
 | |
3BHG
 
 | | Crystal structure of adenylosuccinate lyase from Legionella pneumophila | | 分子名称: | Adenylosuccinate lyase, GLYCEROL, SULFATE ION | | 著者 | Chang, C, Li, H, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-11-28 | | 公開日 | 2007-12-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of adenylosuccinate lyase from Legionella pneumophila.
To be Published
|
|
5YYF
 
 | |
3CBD
 
 | |
3CCY
 
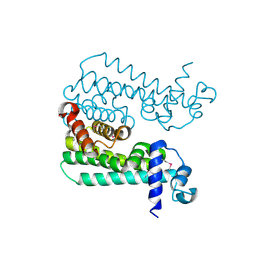 | |
3D3Y
 
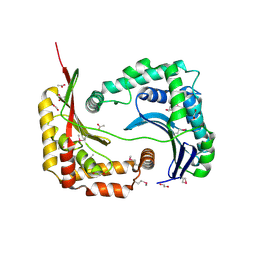 | | Crystal structure of a conserved protein from Enterococcus faecalis V583 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Uncharacterized protein | | 著者 | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-05-13 | | 公開日 | 2008-07-15 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The crystal structure of a conserved protein from Enterococcus faecalis V583.
To be Published
|
|
3D37
 
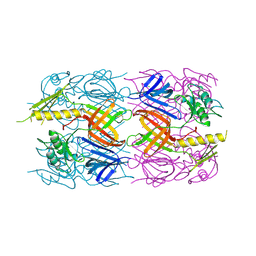 | | The crystal structure of the tail protein from Neisseria meningitidis MC58 | | 分子名称: | CHLORIDE ION, Tail protein, 43 kDa | | 著者 | Zhang, R, Li, H, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-05-09 | | 公開日 | 2008-07-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of the tail protein from Neisseria meningitidis MC58.
To be Published
|
|
3CIT
 
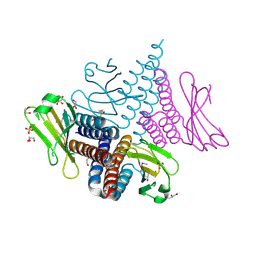 | | Crystal structure of the GAF domain of a putative sensor histidine kinase from Pseudomonas syringae pv. tomato | | 分子名称: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | 著者 | Cuff, M.E, Li, H, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-03-11 | | 公開日 | 2008-05-13 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the GAF domain of a putative sensor histidine kinase from Pseudomonas syringae pv. tomato
To be Published
|
|
7E7C
 
 | |
8GB3
 
 | |
