1F0B
 
 | |
6SJM
 
 | | Crystal structure of the Retinoic Acid Receptor alpha in complex with compound 24 (JP175) | | 分子名称: | 2-[4-[3,5-bis(trifluoromethyl)phenyl]phenyl]ethanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | 著者 | Chaikuad, A, Pollinger, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2019-08-13 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | A Novel Biphenyl-based Chemotype of Retinoid X Receptor Ligands Enables Subtype and Heterodimer Preferences.
Acs Med.Chem.Lett., 10, 2019
|
|
3F03
 
 | |
3FI8
 
 | | Crystal structure of choline kinase from Plasmodium Falciparum, PF14_0020 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Choline kinase, MAGNESIUM ION, ... | | 著者 | Wernimont, A.K, Pizarro, J.C, Artz, J.D, Amaya, M.F, Xiao, T, Lew, J, Wasney, G, Senesterra, G, Kozieradzki, I, Cossar, D, Vedadi, M, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | 登録日 | 2008-12-11 | | 公開日 | 2009-02-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of choline kinase from Plasmodium Falciparum, PF14_0020
TO BE PUBLISHED
|
|
1DQR
 
 | | CRYSTAL STRUCTURE OF RABBIT PHOSPHOGLUCOSE ISOMERASE, A GLYCOLYTIC ENZYME THAT MOONLIGHTS AS NEUROLEUKIN, AUTOCRINE MOTILITY FACTOR, AND DIFFERENTIATION MEDIATOR | | 分子名称: | 6-PHOSPHOGLUCONIC ACID, PHOSPHOGLUCOSE ISOMERASE | | 著者 | Bahnson, B.J, Jeffery, C.J, Ringe, D, Petsko, G.A. | | 登録日 | 2000-01-05 | | 公開日 | 2000-02-09 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of rabbit phosphoglucose isomerase, a glycolytic enzyme that moonlights as neuroleukin, autocrine motility factor, and differentiation mediator.
Biochemistry, 39, 2000
|
|
3F1J
 
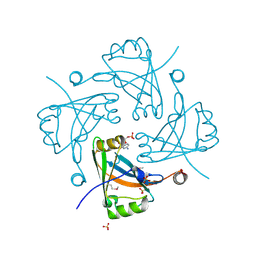 | | Crystal structure of the Borna disease virus matrix protein (BDV-M) reveals RNA binding properties | | 分子名称: | CYTIDINE-5'-MONOPHOSPHATE, Matrix protein, SULFATE ION | | 著者 | Neumann, P, Lieber, D, Meyer, S, Dautel, P, Kerth, A, Kraus, I, Garten, W, Stubbs, M.T. | | 登録日 | 2008-10-28 | | 公開日 | 2009-02-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal structure of the Borna disease virus matrix protein (BDV-M) reveals ssRNA binding properties
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2LV8
 
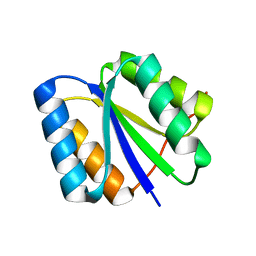 | | Solution NMR Structure de novo designed rossmann 2x2 fold protein, Northeast Structural Genomics Consortium (NESG) Target OR16 | | 分子名称: | De novo designed rossmann 2x2 fold protein | | 著者 | Liu, G, Koga, R, Koga, N, Xiao, R, Pederson, K, Hamilton, K, Ciccosanti, C, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-06-29 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
3VX3
 
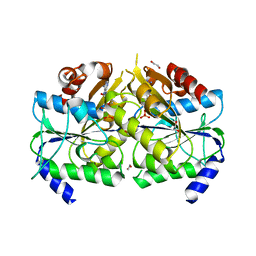 | | Crystal structure of [NiFe] hydrogenase maturation protein HypB from Thermococcus kodakarensis KOD1 | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ATPase involved in chromosome partitioning, ... | | 著者 | Sasaki, D, Watanabe, S, Miki, K. | | 登録日 | 2012-09-09 | | 公開日 | 2013-02-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification and Structure of a Novel Archaeal HypB for [NiFe] Hydrogenase Maturation
J.Mol.Biol., 425, 2013
|
|
6SDR
 
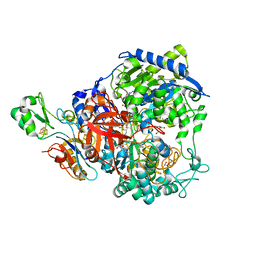 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Oxidized form | | 分子名称: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase, alpha subunit, ... | | 著者 | Oliveira, A.R, Mota, C, Mourato, C, Domingos, R.M, Santos, M.F.A, Gesto, D, Guigliarelli, B, Santos-Silva, T, Romao, M.J, Pereira, I.C. | | 登録日 | 2019-07-29 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Towards the mechanistic understanding of enzymatic CO2 reduction
Acs Catalysis, 2020
|
|
6SE5
 
 | |
7ZS7
 
 | | Crystal structure of human cathepsin L with covalently bound calpain inhibitor VI | | 分子名称: | (2S)-2-[(4-fluorophenyl)sulfonylamino]-3-methyl-N-[(2S)-4-methyl-1-oxidanyl-pentan-2-yl]butanamide, ACETATE ION, Cathepsin L, ... | | 著者 | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | 登録日 | 2022-05-06 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 2024
|
|
6SKL
 
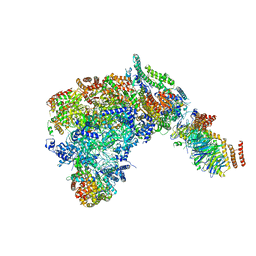 | | Cryo-EM structure of the CMG Fork Protection Complex at a replication fork - Conformation 1 | | 分子名称: | Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA fork, ... | | 著者 | Yeeles, J, Baretic, D, Jenkyn-Bedford, M. | | 登録日 | 2019-08-16 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork.
Mol.Cell, 78, 2020
|
|
2M5Q
 
 | |
3VZ0
 
 | | Structural insights into cofactor and substrate selection by Gox0499 | | 分子名称: | NONAETHYLENE GLYCOL, Putative NAD-dependent aldehyde dehydrogenase | | 著者 | Yuan, Y.A, Yuan, Z, Yin, B, Wei, D. | | 登録日 | 2012-10-09 | | 公開日 | 2013-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for cofactor and substrate selection by cyanobacterium succinic semialdehyde dehydrogenase
J.Struct.Biol., 182, 2013
|
|
3F3U
 
 | | Kinase domain of cSrc in complex with inhibitor RL37 (Type III) | | 分子名称: | 1-[1-(3-aminophenyl)-3-tert-butyl-1H-pyrazol-5-yl]-3-phenylurea, Proto-oncogene tyrosine-protein kinase Src | | 著者 | Gruetter, C, Klueter, S, Getlik, M, Rauh, D. | | 登録日 | 2008-10-31 | | 公開日 | 2009-03-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A new screening assay for allosteric inhibitors of cSrc
Nat.Chem.Biol., 5, 2009
|
|
3W0O
 
 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia, ternary complex with ADP and hygromycin B | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase | | 著者 | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | 登録日 | 2012-11-02 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
2MGW
 
 | | Solution Structure of the UBA Domain of Human NBR1 | | 分子名称: | Next to BRCA1 gene 1 protein | | 著者 | Walinda, E, Morimoto, D, Sugase, K, Komatsu, M, Tochio, H, Shirakawa, M. | | 登録日 | 2013-11-09 | | 公開日 | 2014-04-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the ubiquitin-associated (UBA) domain of human autophagy receptor NBR1 and its interaction with ubiquitin and polyubiquitin.
J.Biol.Chem., 289, 2014
|
|
6SI6
 
 | | N-terminal domain of Drosophila X virus VP3 | | 分子名称: | GLYCEROL, IMIDAZOLE, Structural polyprotein | | 著者 | Ferrero, D.S, Garriga, D, Guerra, P, Uson, I, Verdaguer, N. | | 登録日 | 2019-08-08 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structure and dsRNA-binding activity of the Birnavirus Drosophila X Virus VP3 protein.
J.Virol., 2020
|
|
6SQ2
 
 | |
2LXX
 
 | | Solution structure of cofilin like UNC-60B protein from Caenorhabditis elegans | | 分子名称: | Actin-depolymerizing factor 2, isoform c | | 著者 | Shukla, V, Yadav, R, Kabra, A, Jain, A, Kumar, D, Ono, S, Arora, A. | | 登録日 | 2012-09-04 | | 公開日 | 2013-10-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of UNC-60B from Caenorhabditis elegans
To be Published
|
|
3IW5
 
 | | Human p38 MAP Kinase in Complex with an Indole Derivative | | 分子名称: | Mitogen-activated protein kinase 14, N-[2-(3-{[2-(2,3-dihydro-1,4-benzodioxin-6-ylamino)-2-oxoethyl]sulfanyl}-1H-indol-1-yl)ethyl]-3-(trifluoromethyl)benzamide, octyl beta-D-glucopyranoside | | 著者 | Gruetter, C, Simard, J.R, Rauh, D. | | 登録日 | 2009-09-02 | | 公開日 | 2009-11-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
1DZ1
 
 | | Mouse HP1 (M31) C terminal (shadow chromo) domain | | 分子名称: | MODIFIER 1 PROTEIN | | 著者 | Brasher, S.V, Smith, B.O, Fogh, R.H, Nietlispach, D, Thiru, A, Nielsen, P.R, Broadhurst, R.W, Ball, L.J, Murzina, N, Laue, E.D. | | 登録日 | 2000-02-11 | | 公開日 | 2000-04-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Structure of Mouse Hp1 Suggests a Unique Mode of Single Peptide Recognition by the Shadow Chromo Domain Dimer
Embo J., 19, 2000
|
|
6SKF
 
 | | Cryo-EM Structure of T. kodakarensis 70S ribosome | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Matzov, D, Sas-Chen, A, Thomas, J.M, Santangelo, T, Meier, J.L, Schwartz, S, Shalev-Benami, M. | | 登録日 | 2019-08-15 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping.
Nature, 583, 2020
|
|
6SRW
 
 | | Kemp Eliminase HG3.17 mutant Q50F, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, GLYCEROL, Kemp Eliminase HG3.17 Q50F | | 著者 | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | 登録日 | 2019-09-06 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
2M23
 
 | |
