8IMY
 
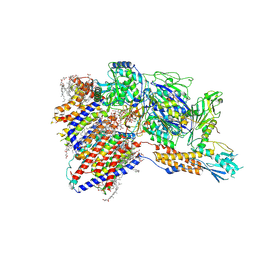 | | Cryo-EM structure of GPI-T (inactive mutant) with GPI and proULBP2, a proprotein substrate | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Li, T, Xu, Y, Qu, Q, Li, D. | | 登録日 | 2023-03-07 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
7CKQ
 
 | |
8H70
 
 | |
7EP0
 
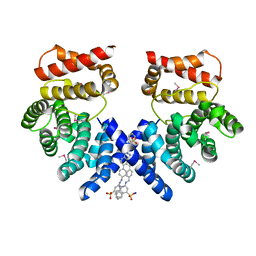 | | Crystal structure of ZYG11B bound to GSTE degron | | 分子名称: | Protein zyg-11 homolog B, sodium 3,3'-(1E,1'E)-biphenyl-4,4'-diylbis(diazene-2,1-diyl)bis(4-aminonaphthalene-1-sulfonate) | | 著者 | Yan, X, Li, Y. | | 登録日 | 2021-04-26 | | 公開日 | 2021-07-14 | | 最終更新日 | 2021-09-01 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP3
 
 | |
7EP2
 
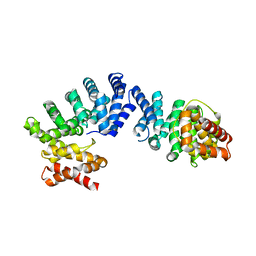 | |
7EP5
 
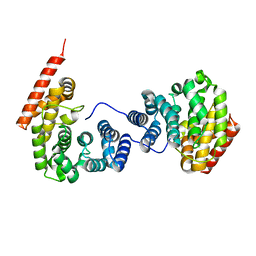 | |
7EP1
 
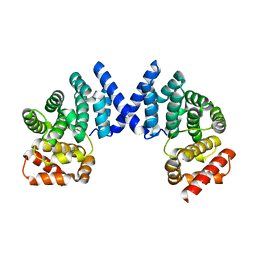 | |
7EP4
 
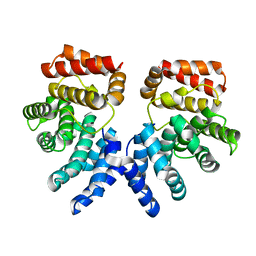 | |
7FIL
 
 | |
7FIK
 
 | | The cryo-EM structure of the CR subunit from X. laevis NPC | | 分子名称: | MGC154553 protein, MGC83295 protein, MGC83926 protein, ... | | 著者 | Shi, Y, Huang, G, Zhan, X. | | 登録日 | 2021-07-31 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex.
Science, 376, 2022
|
|
7C82
 
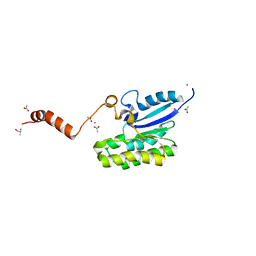 | |
5XGH
 
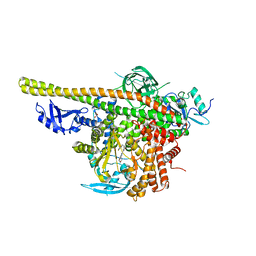 | | Crystal structure of PI3K complex with an inhibitor | | 分子名称: | 3-[(4-fluorophenyl)methylamino]-5-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | 著者 | Song, K, Yang, X, Zhao, Y, Jian, Z. | | 登録日 | 2017-04-13 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.97 Å) | | 主引用文献 | New Insights into PI3K Inhibitor Design using X-ray Structures of PI3K alpha Complexed with a Potent Lead Compound.
Sci Rep, 7, 2017
|
|
7C84
 
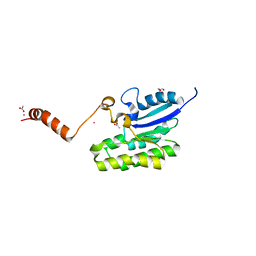 | | Esterase AlinE4 mutant, D162A | | 分子名称: | ACETATE ION, CADMIUM ION, GLYCEROL, ... | | 著者 | Li, Z, Li, J. | | 登録日 | 2020-05-28 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.552 Å) | | 主引用文献 | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
7C85
 
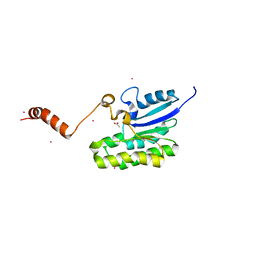 | | Esterase AlinE4 mutant-S13A | | 分子名称: | ACETATE ION, CADMIUM ION, SGNH-hydrolase family esterase | | 著者 | Li, Z, Li, J. | | 登録日 | 2020-05-28 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
7D3U
 
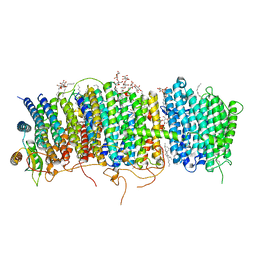 | | Structure of Mrp complex from Dietzia sp. DQ12-45-1b | | 分子名称: | Cation antiporter, DODECYL-BETA-D-MALTOSIDE, Monovalent Na+/H+ antiporter subunit A, ... | | 著者 | Li, B, Zhang, K.D, Wu, X.L, Zhang, X.C. | | 登録日 | 2020-09-21 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of the Dietzia Mrp complex reveals molecular mechanism of this giant bacterial sodium proton pump.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7DTD
 
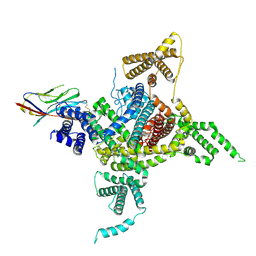 | | Voltage-gated sodium channel Nav1.1 and beta4 | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 1 subunit alpha, ... | | 著者 | Yan, N, Pan, X, Li, Z, Huang, G. | | 登録日 | 2021-01-04 | | 公開日 | 2021-04-07 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Comparative structural analysis of human Na v 1.1 and Na v 1.5 reveals mutational hotspots for sodium channelopathies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7E23
 
 | | SARS-CoV-2 spike in complex with the CA521 neutralizing antibody Fab (focused refinement on Fab-RBD) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CA521 Heavy Chain, CA521 Light Chain, ... | | 著者 | Liu, C, Song, D, Dou, C. | | 登録日 | 2021-02-04 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure and function analysis of a potent human neutralizing antibody CA521 FALA against SARS-CoV-2.
Commun Biol, 4, 2021
|
|
7X5B
 
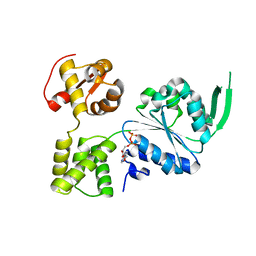 | | Crystal structure of RuvB | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z, Dai, L. | | 登録日 | 2022-03-04 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X5A
 
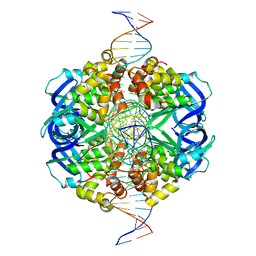 | | CryoEM structure of RuvA-Holliday junction complex | | 分子名称: | DNA (26-MER), Holliday junction ATP-dependent DNA helicase RuvA | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | 登録日 | 2022-03-04 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7P
 
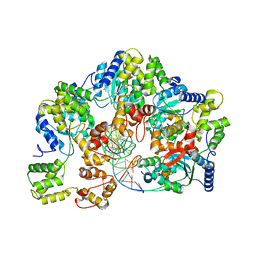 | | CryoEM structure of dsDNA-RuvB-RuvA domain3 complex | | 分子名称: | DNA, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | 登録日 | 2022-03-10 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (7.02 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7Q
 
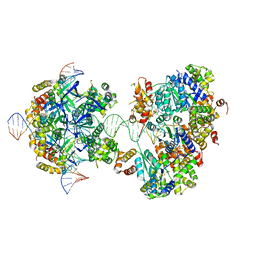 | | CryoEM structure of RuvA-RuvB-Holliday junction complex | | 分子名称: | DNA (26-MER), DNA (40-MER), Holliday junction ATP-dependent DNA helicase RuvA, ... | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | 登録日 | 2022-03-10 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (7.02 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7BAJ
 
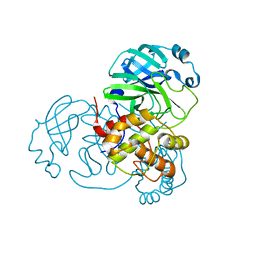 | |
7BAK
 
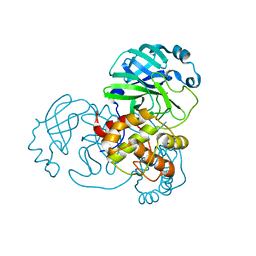 | |
7BAL
 
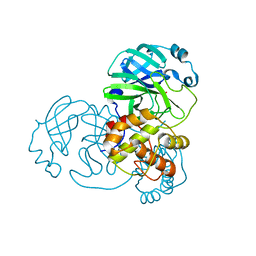 | |
