5FGW
 
 | | Structure of Sda1 nuclease with bound zinc ion | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Moon, A.F, Krahn, J.M, Xun, L, Cuneo, M.J, Pedersen, L.C. | | 登録日 | 2015-12-21 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural characterization of the virulence factor Sda1 nuclease from Streptococcus pyogenes.
Nucleic Acids Res., 44, 2016
|
|
6N52
 
 | | Metabotropic Glutamate Receptor 5 Apo Form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5 | | 著者 | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | 登録日 | 2018-11-20 | | 公開日 | 2019-01-23 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
5FCC
 
 | | Structure of HutD from Pseudomonas fluorescens SBW25 (NaCl condition) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, HutD, ... | | 著者 | Johnston, J.M, Gerth, M.L, Baker, E.N, Lott, J.S, Rainey, P.B. | | 登録日 | 2015-12-15 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structure of HutD from Pseudomonas fluorescens
To Be Published
|
|
6N64
 
 | | Crystal structure of mouse SMCHD1 hinge domain | | 分子名称: | Structural maintenance of chromosomes flexible hinge domain-containing protein 1, Uncharacterized peptide from Structural maintenance of chromosomes flexible hinge domain-containing protein 1 | | 著者 | Birkinshaw, R.W, Chen, K, Czabotar, P.E, Blewitt, M.E, Murphy, J.M. | | 登録日 | 2018-11-25 | | 公開日 | 2020-06-17 | | 最終更新日 | 2021-01-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal structure of the hinge domain of Smchd1 reveals its dimerization mode and nucleic acid-binding residues.
Sci.Signal., 13, 2020
|
|
5FFF
 
 | |
2XU1
 
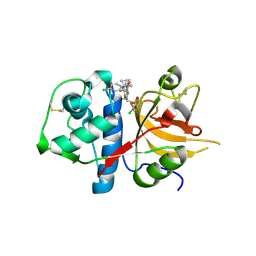 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | 分子名称: | (2S,4R)-1-[1-(4-chlorophenyl)cyclopropyl]carbonyl-4-(2-chlorophenyl)sulfonyl-N-[1-(iminomethyl)cyclopropyl]pyrrolidine-2-carboxamide, CATHEPSIN L1 | | 著者 | Banner, D.W, Benz, J.M, Steinbacher, S, Haap, W. | | 登録日 | 2010-10-14 | | 公開日 | 2011-01-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Systematic Investigation of Halogen Bonding in Protein-Ligand Interactions.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
5FOJ
 
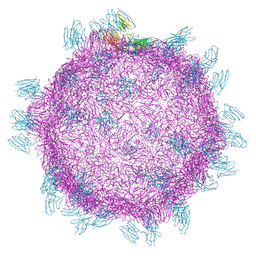 | | Cryo electron microscopy structure of Grapevine Fanleaf Virus complex with Nanobody | | 分子名称: | Nanobody, RNA2 polyprotein | | 著者 | Orlov, I, Hemmer, C, Ackerer, L, Lorber, B, Ghannam, A, Poignavent, V, Hleibieh, K, Sauter, C, Schmitt-Keichinger, C, Belval, L, Hily, J.M, Marmonier, A, Komar, V, Gersch, S, Schellenberger, P, Bron, P, Vigne, E, Muyldermans, S, Lemaire, O, Demangeat, G, Ritzenthaler, C, Klaholz, B.P. | | 登録日 | 2015-11-22 | | 公開日 | 2016-01-20 | | 最終更新日 | 2021-08-11 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis of nanobody recognition of grapevine fanleaf virus and of virus resistance loss.
Proc.Natl.Acad.Sci.USA, 2020
|
|
5D7Z
 
 | |
5GLB
 
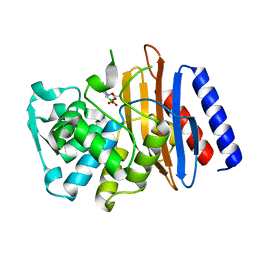 | | Crystal structure of the class A beta-lactamase PenL-tTR10 in complex with CBA | | 分子名称: | Beta-lactamase, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | 著者 | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | 登録日 | 2016-07-10 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5DBU
 
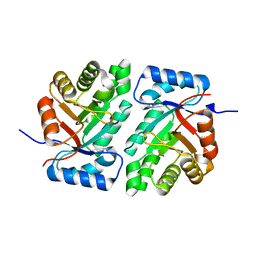 | |
1GGI
 
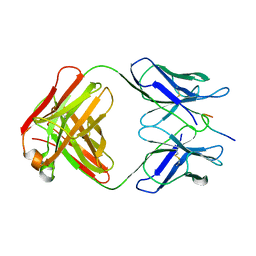 | | CRYSTAL STRUCTURE OF AN HIV-1 NEUTRALIZING ANTIBODY 50.1 IN COMPLEX WITH ITS V3 LOOP PEPTIDE ANTIGEN | | 分子名称: | HIV-1 V3 LOOP PEPTIDE ANTIGEN, IGG2A 50.1 FAB (HEAVY CHAIN), IGG2A 50.1 FAB (LIGHT CHAIN) | | 著者 | Stanfield, R.L, Rini, J.M, Wilson, I.A. | | 登録日 | 1993-04-02 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
5D6W
 
 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A | | 分子名称: | Lysine-specific demethylase 4A, S,R MESO-TARTARIC ACID | | 著者 | Wang, F, Su, Z, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-08-13 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.992 Å) | | 主引用文献 | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
6OEI
 
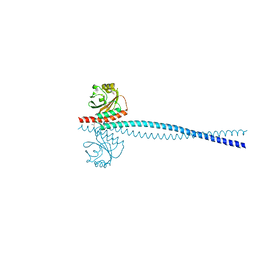 | | Yeast Spc42 N-terminal coiled-coil fused to PDB: 3K2N | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Spindle pole body component SPC42,Sigma-54-dependent transcriptional regulator | | 著者 | Drennan, A.C, Krishna, S, Seeger, M.A, Andreas, M.P, Gardner, J.M, Sether, E.K.R, Jaspersen, S.L, Rayment, I. | | 登録日 | 2019-03-27 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structure and function of Spc42 coiled-coils in yeast centrosome assembly and duplication.
Mol.Biol.Cell, 30, 2019
|
|
6OF2
 
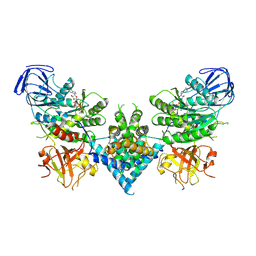 | | Precursor ribosomal RNA processing complex, State 2. | | 分子名称: | CLP1_P domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | 登録日 | 2019-03-28 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OEC
 
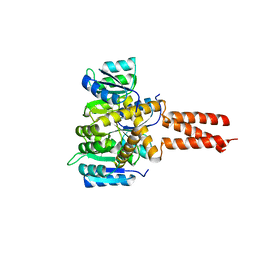 | | Yeast Spc42 Trimeric Coiled-Coil Amino Acids 181-211 fused to PDB: 3H5I | | 分子名称: | CALCIUM ION, Response regulator/sensory box protein/GGDEF domain protein,Spindle pole body component SPC42 | | 著者 | Drennan, A.C, Shivaani, K, Seeger, M.A, Andreas, M.P, Gardner, J.M, Sether, E.K.R, Jasperson, S.L, Rayment, I. | | 登録日 | 2019-03-27 | | 公開日 | 2019-04-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.514 Å) | | 主引用文献 | Structure and function of Spc42 coiled-coils in yeast centrosome assembly and duplication.
Mol.Biol.Cell, 30, 2019
|
|
6OF3
 
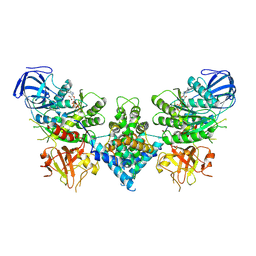 | | Precursor ribosomal RNA processing complex, State 1. | | 分子名称: | CLP1_P domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | 登録日 | 2019-03-28 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5D98
 
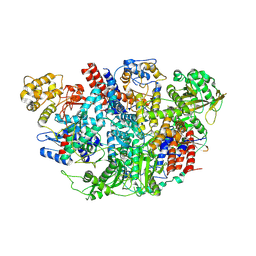 | | Influenza C Virus RNA-dependent RNA Polymerase - Space group P43212 | | 分子名称: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | 著者 | Hengrung, N, El Omari, K, Serna Martin, I, Vreede, F.T, Cusack, S, Rambo, R.P, Vonrhein, C, Bricogne, G, Stuart, D.I, Grimes, J.M, Fodor, E. | | 登録日 | 2015-08-18 | | 公開日 | 2015-10-21 | | 最終更新日 | 2017-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Crystal structure of the RNA-dependent RNA polymerase from influenza C virus.
Nature, 527, 2015
|
|
6OMS
 
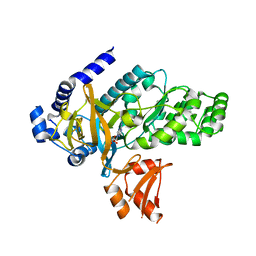 | | Arabidopsis GH3.12 with Chorismate | | 分子名称: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, 4-substituted benzoates-glutamate ligase GH3.12, ADENOSINE MONOPHOSPHATE | | 著者 | Zubieta, C, Westfall, C.S, Holland, C.K, Jez, J.M. | | 登録日 | 2019-04-19 | | 公開日 | 2019-10-09 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.942 Å) | | 主引用文献 | Brassicaceae-specific Gretchen Hagen 3 acyl acid amido synthetases conjugate amino acids to chorismate, a precursor of aromatic amino acids and salicylic acid.
J.Biol.Chem., 294, 2019
|
|
4N6W
 
 | | X-Ray Crystal Structure of Citrate-bound PhnZ | | 分子名称: | CITRATE ANION, FE (III) ION, Predicted HD phosphohydrolase PhnZ | | 著者 | Worsdorfer, B, Lingaraju, M, Yennawar, N.H, Boal, A.K, Krebs, C, Bollinger Jr, J.M, Pandelia, M.E. | | 登録日 | 2013-10-14 | | 公開日 | 2013-11-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Organophosphonate-degrading PhnZ reveals an emerging family of HD domain mixed-valent diiron oxygenases.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6O86
 
 | |
6O9U
 
 | | KirBac3.1 at a resolution of 2 Angstroms | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3,3',3''-phosphoryltripropanoic acid, BARIUM ION, ... | | 著者 | Gulbis, J.M, Clarke, O.B. | | 登録日 | 2019-03-15 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A constricted opening in Kir channels does not impede potassium conduction.
Nat Commun, 11, 2020
|
|
6OK8
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS K127L at cryogenic temperature | | 分子名称: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | 著者 | Jeliazkov, J.R, Robinson, A.C, Berger, J.M, Garcia-Moreno E, B, Gray, J.G. | | 登録日 | 2019-04-12 | | 公開日 | 2019-05-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Toward the computational design of protein crystals with improved resolution.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5CWA
 
 | | Structure of Anthranilate Synthase Component I (TrpE) from Mycobacterium tuberculosis with inhibitor bound | | 分子名称: | 3-{[(1Z)-1-carboxyprop-1-en-1-yl]oxy}-2-hydroxybenzoic acid, Anthranilate synthase component 1, GLYCEROL, ... | | 著者 | Johnston, J.M, Bashiri, G, Evans, G.L, Lott, J.S, Baker, E.N. | | 登録日 | 2015-07-28 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and inhibition of subunit I of the anthranilate synthase complex of Mycobacterium tuberculosis and expression of the active complex.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6OLQ
 
 | | Protein Tyrosine Phosphatase 1B (1-301), P188A mutant, apo state | | 分子名称: | ACETATE ION, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Cui, D.S, Lipchock, J.M, Loria, J.P. | | 登録日 | 2019-04-16 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Uncovering the Molecular Interactions in the Catalytic Loop That Modulate the Conformational Dynamics in Protein Tyrosine Phosphatase 1B.
J.Am.Chem.Soc., 141, 2019
|
|
6NCM
 
 | | Crystal structure of the human FOXN3 DNA binding domain in complex with a forkhead-like (FHL) DNA sequence | | 分子名称: | DNA (5'-D(*AP*TP*AP*GP*CP*GP*TP*CP*TP*TP*AP*GP*CP*AP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*TP*GP*CP*TP*AP*AP*GP*AP*CP*GP*CP*TP*A)-3'), Forkhead box protein N3, ... | | 著者 | Rogers, J.M, Jarrett, S.M, Seegar, T.C, Waters, C.T, Hallworth, A.N, Blacklow, S.C, Bulyk, M.L. | | 登録日 | 2018-12-11 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.704 Å) | | 主引用文献 | Bispecific Forkhead Transcription Factor FoxN3 Recognizes Two Distinct Motifs with Different DNA Shapes.
Mol. Cell, 74, 2019
|
|
