6WSJ
 
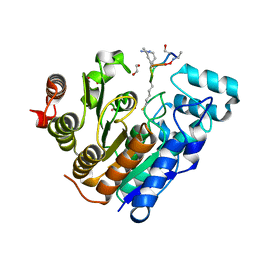 | |
6WHZ
 
 | | Histone deacetylases complex with peptide macrocycles | | 分子名称: | Histone deacetylase 2, SODIUM ION, TETRAETHYLENE GLYCOL, ... | | 著者 | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | 登録日 | 2020-04-08 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
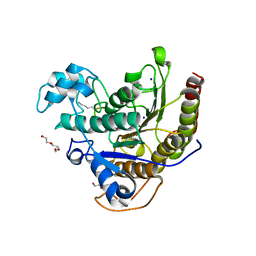
6WI3
 
 | | Histone deacetylases complex with peptide macrocycles | | 分子名称: | (SHA)W(DTH)DN(DSN)(DME)(DAS)K peptide macrocycle, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, ... | | 著者 | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | 登録日 | 2020-04-08 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
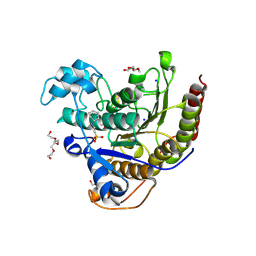
7XR5
 
 | | Crystal structure of imine reductase with NAPDH from Streptomyces albidoflavus | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45,48,51,54,57-nonadecaoxanonapentacontane-1,59-diol, 6-phosphogluconate dehydrogenase NAD-binding, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zhang, J, Chen, R.C, Gao, S.S. | | 登録日 | 2022-05-09 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Actinomycetes-derived imine reductases with a preference towards bulky amine substrates.
Commun Chem, 5, 2022
|
|
3EZX
 
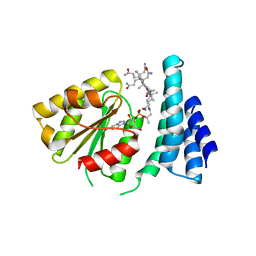 | |
3CQ0
 
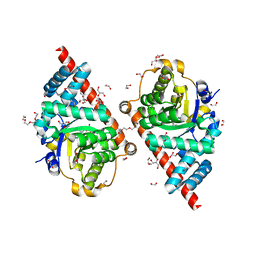 | | Crystal Structure of TAL2_YEAST | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Putative transaldolase YGR043C, ... | | 著者 | Huang, H, Niu, L, Teng, M. | | 登録日 | 2008-04-01 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The crystal structure and identification of NQM1/YGR043C, a transaldolase from Saccharomyces cerevisiae
Proteins, 73, 2008
|
|
8HLP
 
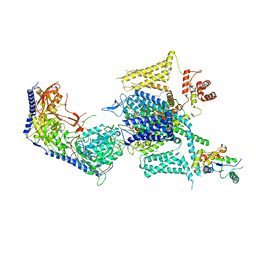 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 (apo) | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Wei, Y, Yu, Z, Zhao, Y. | | 登録日 | 2022-11-30 | | 公開日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
8HMB
 
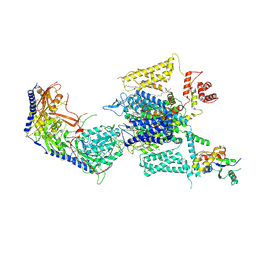 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with benidipine (BEN) | | 分子名称: | (3R)-1-benzylpiperidin-3-yl methyl (2R,3R,4R,5R,6S)-2,6-dimethyl-4-(3-nitrophenyl)piperidine-3,5-dicarboxylate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wei, Y, Yu, Z, Zhao, Y. | | 登録日 | 2022-12-02 | | 公開日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
8HMA
 
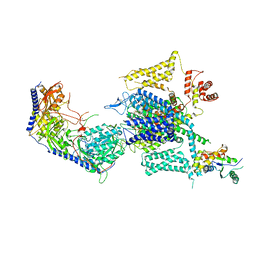 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with tetrandrine (TET) | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6,6',7,12-tetramethoxy-2,2'-dimethyl-1beta-3,4-didehydroberbaman, ... | | 著者 | Wei, Y, Yu, Z, Zhao, Y. | | 登録日 | 2022-12-02 | | 公開日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
7E7X
 
 | |
7E7Y
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | 分子名称: | 368-2 H, 368-2 L, 604 H, ... | | 著者 | Du, S, Xiao, J, Zhang, Z. | | 登録日 | 2021-03-01 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E88
 
 | |
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | 分子名称: | 368-2 H, 368-2 L, 604 H, ... | | 著者 | Du, S, Xiao, J, Zhang, Z. | | 登録日 | 2021-03-01 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E86
 
 | |
8HR2
 
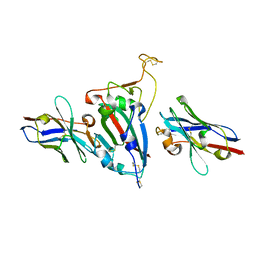 | |
8PO0
 
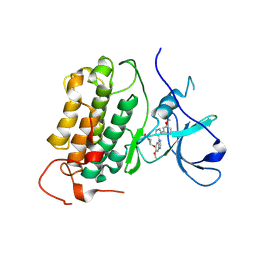 | |
8PO1
 
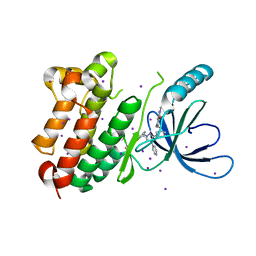 | |
8PO4
 
 | |
8PO3
 
 | |
8PO2
 
 | |
8PNZ
 
 | |
3FW5
 
 | |
3FW4
 
 | |
7KGB
 
 | |
