8IPK
 
 | |
8IPL
 
 | |
8IPM
 
 | |
8J6Z
 
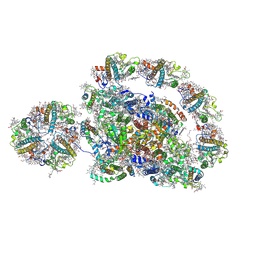 | | Cryo-EM structure of the Arabidopsis thaliana photosystem I(PSI-LHCII-ST2) | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Chen, S.J.B, Wu, J.H, Sui, S.F, Zhang, L.X. | | 登録日 | 2023-04-26 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.79 Å) | | 主引用文献 | Regulatory dynamics of the higher-plant PSI-LHCI supercomplex during state transitions.
Mol Plant, 16, 2023
|
|
8J7A
 
 | | Coordinates of Cryo-EM structure of the Arabidopsis thaliana PSI in state 1 (PSI-ST1) | | 分子名称: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | 著者 | Chen, S.J.B, Wu, J.H, Sui, S.F, Zhang, L.X. | | 登録日 | 2023-04-27 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Regulatory dynamics of the higher-plant PSI-LHCI supercomplex during state transitions.
Mol Plant, 16, 2023
|
|
8J7B
 
 | | Coordinates of Cryo-EM structure of the Arabidopsis thaliana PSI in state 2 (PSI-ST2) | | 分子名称: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | 著者 | Chen, S.J.B, Wu, J.H, Sui, S.F, Zhang, L.X. | | 登録日 | 2023-04-27 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Regulatory dynamics of the higher-plant PSI-LHCI supercomplex during state transitions.
Mol Plant, 16, 2023
|
|
7X2A
 
 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class1 (1u2d RBD with 1Fab) | | 分子名称: | MERS-CoV Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | 著者 | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | 登録日 | 2022-02-25 | | 公開日 | 2022-11-09 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (2.49 Å) | | 主引用文献 | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X26
 
 | | S41 neutralizing antibody Fab(MERS-CoV) | | 分子名称: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | 著者 | Zeng, J.W, Zhang, S.Y, Wang, X.W. | | 登録日 | 2022-02-25 | | 公開日 | 2022-11-09 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.685 Å) | | 主引用文献 | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X29
 
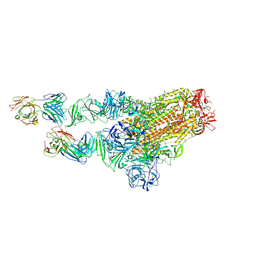 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class2 (1u2d RBD with 2Fab) | | 分子名称: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | 著者 | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | 登録日 | 2022-02-25 | | 公開日 | 2022-11-09 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (2.49 Å) | | 主引用文献 | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X28
 
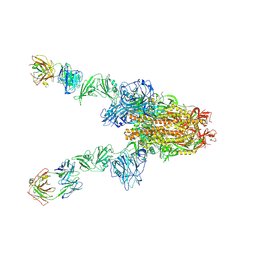 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class3 (2u1d RBD with 2Fab) | | 分子名称: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | 著者 | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | 登録日 | 2022-02-25 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-01-25 | | 実験手法 | ELECTRON MICROSCOPY (2.49 Å) | | 主引用文献 | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7D14
 
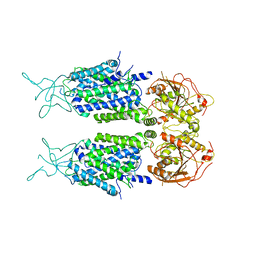 | | Mouse KCC2 | | 分子名称: | Solute carrier family 12 member 5 | | 著者 | Zhang, S, Yang, M. | | 登録日 | 2020-09-13 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2.
Commun Biol, 4, 2021
|
|
7D10
 
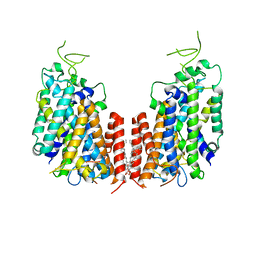 | | Human NKCC1 | | 分子名称: | PALMITIC ACID, Solute carrier family 12 member 2 | | 著者 | Zhang, S, Yang, M. | | 登録日 | 2020-09-12 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | The structural basis of function and regulation of neuronal cotransporters NKCC1 and KCC2.
Commun Biol, 4, 2021
|
|
7CL0
 
 | | Crystal structure of human SIRT6 | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-6, ... | | 著者 | Song, K, Zhang, J. | | 登録日 | 2020-07-20 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
7CL1
 
 | |
7DFG
 
 | |
7DFH
 
 | |
7X25
 
 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class4 (2u1d RBD with 3Fab) | | 分子名称: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | 著者 | Zeng, J, Zhang, S, Zhou, H, Wang, X. | | 登録日 | 2022-02-25 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (2.49 Å) | | 主引用文献 | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7DOK
 
 | |
7DOI
 
 | |
7EKL
 
 | | Mitochondrial outer membrane protein | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family B member 6, MAGNESIUM ION | | 著者 | Zhang, S.S. | | 登録日 | 2021-04-05 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Molecular insights into the human ABCB6 transporter.
Cell Discov, 7, 2021
|
|
3RX3
 
 | | Crystal Structure of Human Aldose Reductase Complexed with Sulindac | | 分子名称: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [(1Z)-5-fluoro-2-methyl-1-{4-[methylsulfinyl]benzylidene}-1H-inden-3-yl]acetic acid | | 著者 | Zheng, X, Chen, J, Luo, H, Hu, X. | | 登録日 | 2011-05-10 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The molecular basis for inhibition of sulindac and its metabolites towards human aldose reductase.
Febs Lett., 586, 2012
|
|
3SO4
 
 | |
3RX4
 
 | | Crystal Structure of Human Aldose Reductase complexed with Sulindac Sulfide | | 分子名称: | 2-[(3Z)-6-fluoranyl-2-methyl-3-[(4-methylsulfanylphenyl)methylidene]inden-1-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zheng, X, Chen, J, Luo, H, Hu, X. | | 登録日 | 2011-05-10 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The molecular basis for inhibition of sulindac and its metabolites towards human aldose reductase.
Febs Lett., 586, 2012
|
|
3RX2
 
 | | Crystal Structure of Human Aldose Reductase Complexed with Sulindac Sulfone | | 分子名称: | 2-[(3Z)-6-fluoranyl-2-methyl-3-[(4-methylsulfonylphenyl)methylidene]inden-1-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zheng, X, Chen, J, Luo, H, Hu, X. | | 登録日 | 2011-05-10 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The molecular basis for inhibition of sulindac and its metabolites towards human aldose reductase
Febs Lett., 586, 2012
|
|
8IEJ
 
 | | RNF20-RNF40/hRad6A-Ub/nucleosome complex | | 分子名称: | DNA (147-MER), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | 著者 | Ai, H, Deng, Z, Sun, M, Du, Y, Pan, M, Liu, L. | | 登録日 | 2023-02-15 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Mechanistic insights into nucleosomal H2B monoubiquitylation mediated by yeast Bre1-Rad6 and its human homolog RNF20/RNF40-hRAD6A.
Mol.Cell, 83, 2023
|
|
