1T8R
 
 | |
4FD3
 
 | |
4G7O
 
 | |
4G7Z
 
 | |
4G7H
 
 | |
3LC8
 
 | |
4Z69
 
 | |
3LBS
 
 | |
3LZC
 
 | | Crystal structure of Dph2 from Pyrococcus horikoshii | | 分子名称: | Dph2 | | 著者 | Zhang, Y, Zhu, X, Torelli, A.T, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | 登録日 | 2010-03-01 | | 公開日 | 2010-06-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.261 Å) | | 主引用文献 | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
4OIN
 
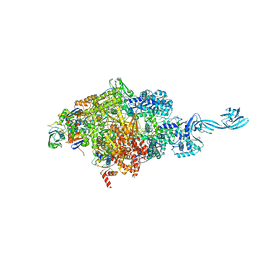 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 | | 分子名称: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | 著者 | Zhang, Y, Ebright, R.H, Arnold, E. | | 登録日 | 2014-01-20 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIO
 
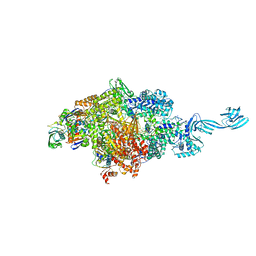 | | Crystal structure of Thermus thermophilus pre-insertion substrate complex for de novo transcription initiation | | 分子名称: | 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ... | | 著者 | Zhang, Y, Ebright, R.H, Arnold, E. | | 登録日 | 2014-01-20 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIP
 
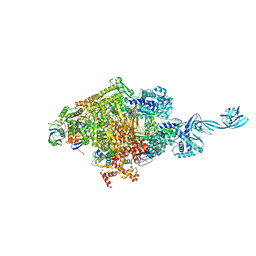 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077, ATP, and CMPcPP | | 分子名称: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | 著者 | Zhang, Y, Ebright, R.H, Arnold, E. | | 登録日 | 2014-01-20 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIQ
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 and rifampicin | | 分子名称: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | 著者 | Zhang, Y, Ebright, R.H, Arnold, E. | | 登録日 | 2014-01-20 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.624 Å) | | 主引用文献 | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIR
 
 | | Crystal structure of Thermus thermophilus RNA polymerase transcription initiation complex soaked with GE23077 and rifamycin SV | | 分子名称: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | 著者 | Zhang, Y, Ebright, R.H, Arnold, E. | | 登録日 | 2014-01-20 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.105 Å) | | 主引用文献 | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
3PQF
 
 | |
3PQE
 
 | |
3PME
 
 | | Crystal structure of the receptor binding domain of botulinum neurotoxin C/D mosaic serotype | | 分子名称: | GLYCEROL, SULFATE ION, Type C neurotoxin | | 著者 | Zhang, Y, Buchko, G.W, Qin, L, Robinson, H, Varnum, S.M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2010-11-16 | | 公開日 | 2010-12-15 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Crystal structure of the receptor binding domain of the botulinum C-D mosaic neurotoxin reveals potential roles of lysines 1118 and 1136 in membrane interactions.
Biochem.Biophys.Res.Commun., 404, 2011
|
|
3PQD
 
 | |
4F83
 
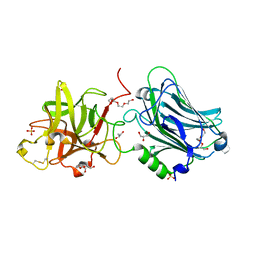 | | Crystal structure of the receptor binding domain of botulinum neurotoxin mosaic serotype C/D with a tetraethylene glycol molecule bound on the Hcn sub-domain and a sulfate ion at the putative active site | | 分子名称: | GLYCEROL, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | 著者 | Zhang, Y, Buchko, G.W, Gardberg, A, Edwards, T.E, Sankaran, B, Robinson, H, Varnum, S.M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2012-05-16 | | 公開日 | 2012-06-20 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural insights into the functional role of the Hcn sub-domain of the receptor-binding domain of the botulinum neurotoxin mosaic serotype C/D.
Biochimie, 95, 2013
|
|
4GKU
 
 | |
5M5G
 
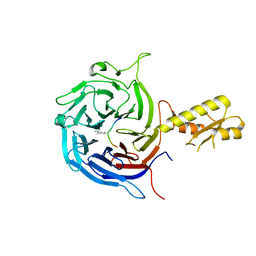 | | Crystal structure of the Chaetomium Thermophilum polycomb repressive complex 2 (PRC2) | | 分子名称: | Fragment from molecular 2 (region containing putative polycomb protein Suz12), HISTONE H3 11-Mer peptide, Putative uncharacterized protein, ... | | 著者 | Zhang, Y, Justin, N, Wilson, J, Gamblin, S. | | 登録日 | 2016-10-21 | | 公開日 | 2017-01-11 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Comment on "Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2".
Science, 354, 2016
|
|
4NPT
 
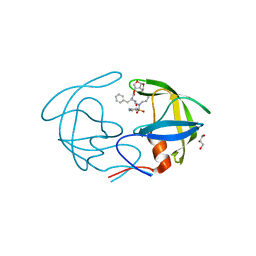 | | Crystal Structure of HIV-1 Protease Multiple Mutant P51 Complexed with Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, GLYCEROL, Protease | | 著者 | Zhang, Y, Weber, I.T. | | 登録日 | 2013-11-22 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Structures of darunavir-resistant HIV-1 protease mutant reveal atypical binding of darunavir to wide open flaps.
Acs Chem.Biol., 9, 2014
|
|
4NPU
 
 | |
4OXW
 
 | |
7ST4
 
 | | Calcium-saturated jGCaMP8.410.80 | | 分子名称: | CALCIUM ION, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Zhang, Y, Looger, L.L. | | 登録日 | 2021-11-11 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Fast and sensitive GCaMP calcium indicators for imaging neural populations
Nature, 615, 2023
|
|
