8CBD
 
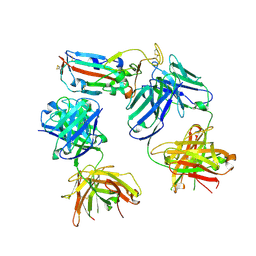 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-1 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-1 heavy chain, BA.4/5-1 light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2023-01-25 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.52 Å) | | 主引用文献 | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8CMA
 
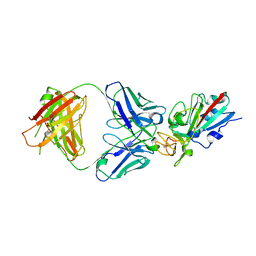 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-35 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-35 heavy chain, BA.4/5-35 light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2023-02-18 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
7ZXU
 
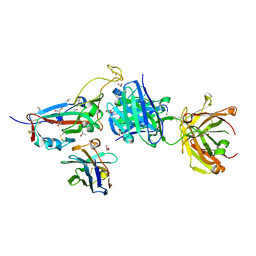 | | SARS-CoV-2 Omicron BA.4/5 RBD in complex with Beta-27 Fab and C1 nanobody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-27 heavy chain, Beta-27 light chain, ... | | 著者 | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2022-05-23 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum.
Cell, 185, 2022
|
|
8BH5
 
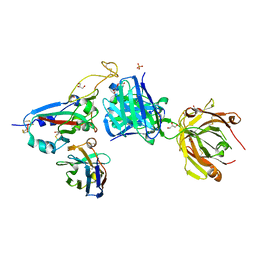 | | SARS-CoV-2 BA.2.12.1 RBD in complex with Beta-27 Fab and C1 nanobody | | 分子名称: | Beta-27 heavy chain, Beta-27 light chain, GLYCEROL, ... | | 著者 | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2022-10-29 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Humoral responses against SARS-CoV-2 Omicron BA.2.11, BA.2.12.1 and BA.2.13 from vaccine and BA.1 serum.
Cell Discov, 8, 2022
|
|
6SNW
 
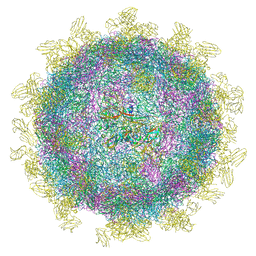 | | Structure of Coxsackievirus A10 complexed with its receptor KREMEN1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP3, ... | | 著者 | Zhao, Y, Zhou, D, Ni, T, Karia, D, Kotecha, A, Wang, X, Rao, Z, Jones, E.Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2019-08-27 | | 公開日 | 2020-01-15 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of Coxsackie Virus A10.
Nat Commun, 11, 2020
|
|
6SNB
 
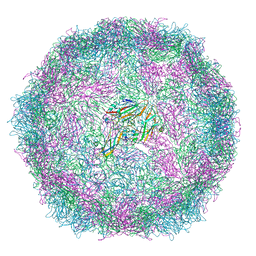 | | Structure of Coxsackievirus A10 A-particle | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | 著者 | Zhao, Y, Zhou, D, Ni, T, Karia, D, Kotecha, A, Wang, X, Rao, Z, Jones, E.Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2019-08-23 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of Coxsackie Virus A10.
Nat Commun, 11, 2020
|
|
6SMG
 
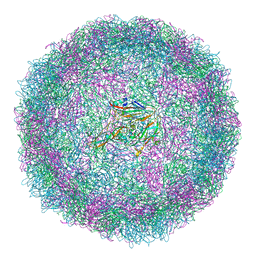 | | Structure of Coxsackievirus A10 | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Zhao, Y, Zhou, D, Ni, T, Karia, D, Kotecha, A, Wang, X, Rao, Z, Jones, E.Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2019-08-21 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of Coxsackie Virus A10.
Nat Commun, 11, 2020
|
|
5O9J
 
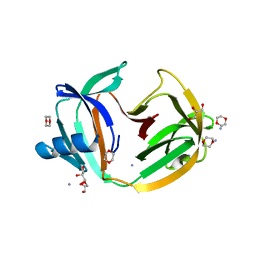 | | Crystal structure of transcription factor IIB Mja mini-intein | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AMMONIUM ION, ... | | 著者 | Mikula, K.M, Iwai, H, Zhou, D, Wlodawer, A. | | 登録日 | 2017-06-19 | | 公開日 | 2017-11-01 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for the Persistence of Homing Endonucleases in Transcription Factor IIB Inteins.
J. Mol. Biol., 429, 2017
|
|
6YM0
 
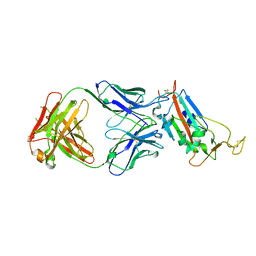 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with CR3022 Fab (crystal form 1) | | 分子名称: | Spike glycoprotein, heavy chain, light chain | | 著者 | Huo, J, Zhao, Y, Ren, J, Zhou, D, Ginn, H.M, Fry, E.E, Owens, R, Stuart, D.I. | | 登録日 | 2020-04-07 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (4.36 Å) | | 主引用文献 | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6YOR
 
 | | Structure of the SARS-CoV-2 spike S1 protein in complex with CR3022 Fab | | 分子名称: | IgG H chain, IgG L chain, Spike glycoprotein | | 著者 | Huo, J, Zhao, Y, Ren, J, Zhou, D, Duyvesteyn, H.M.E, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | 登録日 | 2020-04-15 | | 公開日 | 2020-04-29 | | 最終更新日 | 2022-04-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6YLA
 
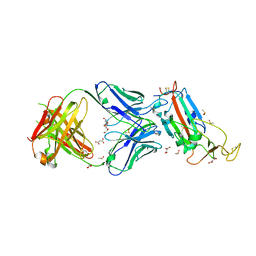 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with CR3022 Fab | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Huo, J, Zhao, Y, Ren, J, Zhou, D, Ginn, H.M, Fry, E.E, Owens, R, Stuart, D.I. | | 登録日 | 2020-04-06 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6Z97
 
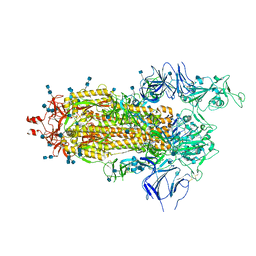 | | Structure of the prefusion SARS-CoV-2 spike glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | 著者 | Duyvesteyn, H.M.E, Ren, J, Zhao, Y, Zhou, D, Huo, J, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | 登録日 | 2020-06-03 | | 公開日 | 2020-07-01 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6Z43
 
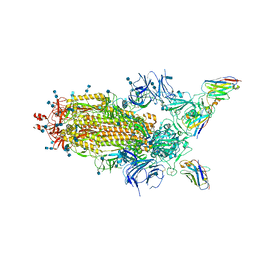 | | Cryo-EM Structure of SARS-CoV-2 Spike : H11-D4 Nanobody Complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, ... | | 著者 | Ruza, R.R, Duyvesteyn, H.M.E, Shah, P, Carrique, L, Ren, J, Malinauskas, T, Zhou, D, Stuart, D.I, Naismith, J.H. | | 登録日 | 2020-05-22 | | 公開日 | 2020-06-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for a potent neutralising single-domain antibody that blocks SARS-CoV-2 binding to its receptor ACE2
To Be Published
|
|
3B4M
 
 | |
8FY0
 
 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-xL) | | 分子名称: | Bcl-2-like protein 1, CACODYLIC ACID, Elongin-B, ... | | 著者 | Olsen, S.K, Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D. | | 登録日 | 2023-01-25 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
8FY2
 
 | | E3:PROTAC:target ternary complex structure (VCB/WH244/BCL-2) | | 分子名称: | Apoptosis regulator Bcl-2, Elongin-B, Elongin-C, ... | | 著者 | Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Ruben, E, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D, Olsen, S.K. | | 登録日 | 2023-01-25 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
8FY1
 
 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-2) | | 分子名称: | Apoptosis regulator Bcl-2, Elongin-B, Elongin-C, ... | | 著者 | Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D, Olsen, S.K. | | 登録日 | 2023-01-25 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
6RVR
 
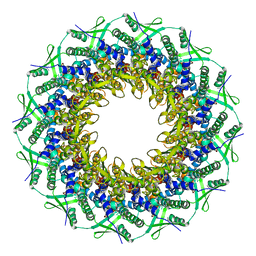 | | Atomic structure of the Epstein-Barr portal, structure I | | 分子名称: | Portal protein | | 著者 | Machon, C, Fabrega-Ferrer, M, Zhou, D, Cuervo, A, Carrascosa, J.L, Stuart, D.I, Coll, M. | | 登録日 | 2019-05-31 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Atomic structure of the Epstein-Barr virus portal.
Nat Commun, 10, 2019
|
|
6RVS
 
 | | Atomic structure of the Epstein-Barr portal, structure II | | 分子名称: | Portal protein | | 著者 | Machon, C, Fabrega-Ferrer, M, Zhou, D, Cuervo, A, Carrascosa, J.L, Stuart, D.I, Coll, M. | | 登録日 | 2019-05-31 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Atomic structure of the Epstein-Barr virus portal.
Nat Commun, 10, 2019
|
|
1Z5L
 
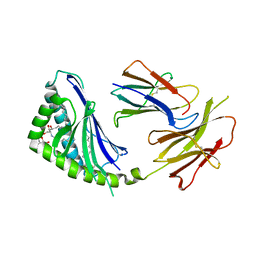 | | Structure of a highly potent short-chain galactosyl ceramide agonist bound to CD1D | | 分子名称: | (2S,3S,4R)-N-OCTANOYL-1-[(ALPHA-D-GALACTOPYRANOSYL)OXY]-2-AMINO-OCTADECANE-3,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zajonc, D.M, Cantu, C, Mattner, J, Zhou, D, Savage, P.B, Bendelac, A, Wilson, I.A, Teyton, L. | | 登録日 | 2005-03-18 | | 公開日 | 2005-07-19 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor.
Nat.Immunol., 6, 2005
|
|
4E2T
 
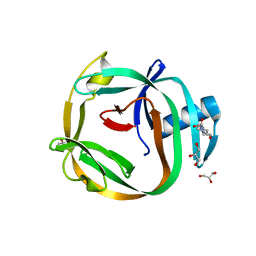 | | Crystal Structures of RadA intein from Pyrococcus horikoshii | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Pho radA intein | | 著者 | Oeemig, J.S, Zhou, D, Kajander, T, Wlodawer, A, Iwai, H. | | 登録日 | 2012-03-09 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | NMR and Crystal Structures of the Pyrococcus horikoshii RadA Intein Guide a Strategy for Engineering a Highly Efficient and Promiscuous Intein.
J.Mol.Biol., 421, 2012
|
|
4E2U
 
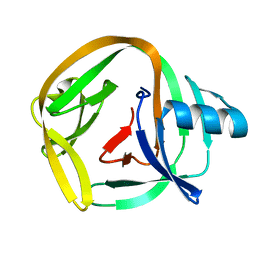 | | Crystal Structures of RadAmin intein from Pyrococcus horikoshii | | 分子名称: | Pho radA intein | | 著者 | Oeemig, J.S, Zhou, D, Kajander, T, Wlodawer, A, Iwai, H. | | 登録日 | 2012-03-09 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.582 Å) | | 主引用文献 | NMR and Crystal Structures of the Pyrococcus horikoshii RadA Intein Guide a Strategy for Engineering a Highly Efficient and Promiscuous Intein.
J.Mol.Biol., 421, 2012
|
|
3NR6
 
 | | Crystal structure of xenotropic murine leukemia virus-related virus (XMRV) protease | | 分子名称: | PHOSPHATE ION, POTASSIUM ION, Protease p14 | | 著者 | Lubkowski, J, Li, M, Gustchina, A, Zhou, D, Dauter, Z, Wlodawer, A. | | 登録日 | 2010-06-30 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Crystal structure of XMRV protease differs from the structures of other retropepsins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2LQM
 
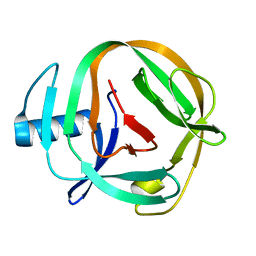 | | Solution Structures of RadA intein from Pyrococcus horikoshii | | 分子名称: | Pho radA intein | | 著者 | Oeemig, J.S, Zhou, D, Kajander, T, Wlodawer, A, Iwai, H. | | 登録日 | 2012-03-09 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR and Crystal Structures of the Pyrococcus horikoshii RadA Intein Guide a Strategy for Engineering a Highly Efficient and Promiscuous Intein.
J.Mol.Biol., 421, 2012
|
|
6BXJ
 
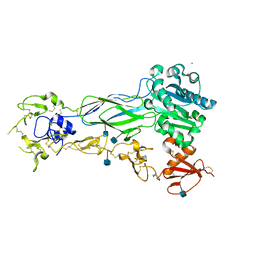 | | Structure of a single-chain beta3 integrin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chimera protein of Integrin beta-3 and Integrin alpha-L, MAGNESIUM ION | | 著者 | Thinn, A.M.M, Wang, Z, Zhou, D, Zhao, Y, Curtis, B.R, Zhu, J. | | 登録日 | 2017-12-18 | | 公開日 | 2018-10-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.092 Å) | | 主引用文献 | Autonomous conformational regulation of beta3integrin and the conformation-dependent property of HPA-1a alloantibodies.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
