6V18
 
 | | immune receptor complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta, GLYCEROL, ... | | 著者 | Lim, J.J, Rossjohn, J. | | 登録日 | 2019-11-20 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR.
Sci Immunol, 6, 2021
|
|
8OKB
 
 | | SARS-CoV2 NSP5 in complex with a peptidomimetic ligand | | 分子名称: | 3C-like proteinase nsp5, methyl (4~{S})-4-[[(2~{S})-4-methyl-2-(phenylmethoxycarbonylamino)pentanoyl]amino]-5-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | 著者 | Calderone, V. | | 登録日 | 2023-03-28 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Development of a GC-376 Based Peptidomimetic PROTAC as a Degrader of 3-Chymotrypsin-like Protease of SARS-CoV-2.
Acs Med.Chem.Lett., 15, 2024
|
|
8OKC
 
 | | SARS-CoV2 NSP5 in complex with a GC-376 based peptidomimetic PROTAC | | 分子名称: | (phenylmethyl) ~{N}-[(2~{R})-1-[[(~{Z},2~{S})-5-[4-[[1-[2-[(3~{R})-2,6-bis(oxidanylidene)piperidin-3-yl]-6-fluoranyl-1,3-bis(oxidanylidene)isoindol-5-yl]piperidin-4-yl]methyl]piperazin-1-yl]-5-oxidanylidene-1-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]pent-3-en-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase nsp5 | | 著者 | Calderone, V. | | 登録日 | 2023-03-28 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Development of a GC-376 Based Peptidomimetic PROTAC as a Degrader of 3-Chymotrypsin-like Protease of SARS-CoV-2.
Acs Med.Chem.Lett., 15, 2024
|
|
7Z62
 
 | | Structure of the LecA lectin from Pseudomonas aeruginosa in complex with a biaryl-thiogalactoside | | 分子名称: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-6-[4-[1-(4-methoxyphenyl)ethenyl]phenyl]sulfanyl-oxane-3,4,5-triol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | 著者 | Varrot, A. | | 登録日 | 2022-03-10 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Discovery of potent 1,1-diarylthiogalactoside glycomimetic inhibitors of Pseudomonas aeruginosa LecA with antibiofilm properties.
Eur.J.Med.Chem., 247, 2022
|
|
7Z63
 
 | |
5MEM
 
 | | A potent fluorescent inhibitor of glycogen phosphorylase as a catalytic site probe. | | 分子名称: | 2-[[1-[(2~{R},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]-2-oxidanylidene-pyrimidin-4-yl]amino]-10~{H}-acridin-9-one, Glycogen phosphorylase, muscle form, ... | | 著者 | Mamais, M, Chrysina, E.D. | | 登録日 | 2016-11-15 | | 公開日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | A New Potent Inhibitor of Glycogen Phosphorylase Reveals the Basicity of the Catalytic Site.
Chemistry, 23, 2017
|
|
7K3G
 
 | | SARS-CoV-2 Envelope Protein Transmembrane Domain: Pentameric Structure Determined by Solid-State NMR | | 分子名称: | Envelope small membrane protein | | 著者 | Mandala, V.S, Hong, M, McKay, M.J, Shcherbakov, A.S, Dregni, A.J. | | 登録日 | 2020-09-11 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4JMP
 
 | | Crystal structure of the chimerical protein CapA2B2 | | 分子名称: | C-terminal fragment of CapA, Protein tyrosine kinase | | 著者 | Olivares-Illana, V, Morera, S, Grangeasse, C, Nessler, S. | | 登録日 | 2013-03-14 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Comparative analysis of the Tyr-kinases CapB1 and CapB2 fused to their cognate modulators CapA1 and CapA2 from Staphylococcus aureus
Plos One, 8, 2013
|
|
6YVK
 
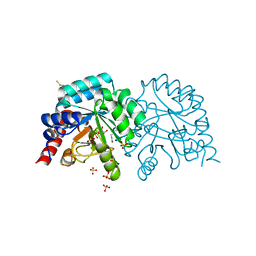 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 0.71 MGy exposure | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-04-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YVL
 
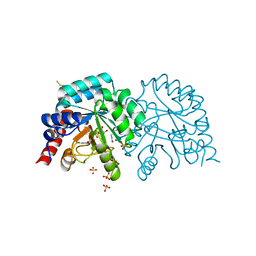 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 1.42 MGy exposure | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-04-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YVM
 
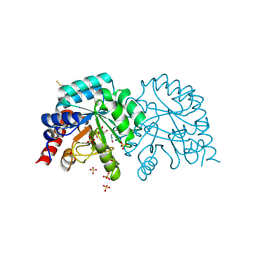 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 2.13 MGy exposure | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-04-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YVN
 
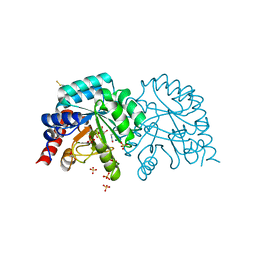 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 2.84 MGy exposure | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-04-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YWT
 
 | |
6YVO
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 3.55 MGy exposure | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-04-28 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6YWU
 
 | | Human OMPD-domain of UMPS (K314AcK) in complex with UMP at 1.1 Angstroms resolution | | 分子名称: | GLYCEROL, SULFATE ION, URIDINE-5'-MONOPHOSPHATE, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-04-30 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6ZWZ
 
 | |
6ZWY
 
 | |
6ZX1
 
 | | OMPD-domain of human UMPS in complex with 6-Aza-UMP at 1.0 Angstroms resolution | | 分子名称: | 6-AZA URIDINE 5'-MONOPHOSPHATE, PROLINE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-07-29 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6ZX3
 
 | | OMPD-domain of human UMPS in complex with 6-thiocarboxamido-UMP at 1.15 Angstroms resolution | | 分子名称: | GLYCEROL, PROLINE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Schimdt, T. | | 登録日 | 2020-07-29 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6ZX2
 
 | | OMPD-domain of human UMPS in complex with 6-carboxamido-UMP at 1.2 Angstroms resolution | | 分子名称: | PROLINE, SULFATE ION, Uridine 5'-monophosphate synthase, ... | | 著者 | Tittmann, K, Rindfleisch, S, Schimdt, T. | | 登録日 | 2020-07-29 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6ZX0
 
 | | OMPD-domain of human UMPS in complex with the substrate OMP at 1.25 Angstroms resolution | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-07-29 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6FO5
 
 | | FIRST DOMAIN OF HUMAN BROMODOMAIN BRD4 IN COMPLEX WITH INHIBITOR #17 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-[[4-[[7-ethyl-2,6-bis(oxidanylidene)purin-3-yl]methyl]phenyl]methyl]-2-oxidanylidene-1,3,4,5-tetrahydro-1-benzazepine-7-sulfonamide | | 著者 | Raux, B, Betzi, S. | | 登録日 | 2018-02-06 | | 公開日 | 2018-06-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Integrated Strategy for Lead Optimization Based on Fragment Growing: The Diversity-Oriented-Target-Focused-Synthesis Approach.
J. Med. Chem., 61, 2018
|
|
7CO1
 
 | | Crystal structure of SMAD2 in complex with wild-type CBP | | 分子名称: | CREB-binding protein, Mothers against decapentaplegic homolog 2 | | 著者 | Miyazono, K, Wada, H, Ito, T, Tanokura, M. | | 登録日 | 2020-08-03 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural basis for transcriptional coactivator recognition by SMAD2 in TGF-beta signaling.
Sci.Signal., 13, 2020
|
|
6FNX
 
 | |
3LJE
 
 | | The X-ray structure of zebrafish RNase5 | | 分子名称: | ACETATE ION, SULFATE ION, Zebrafish RNase5 | | 著者 | Russo Krauss, I, Merlino, A, Coscia, F, Mazzarella, L, Sica, F. | | 登録日 | 2010-01-26 | | 公開日 | 2010-11-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A new RNase sheds light on the RNase/angiogenin subfamily from zebrafish.
Biochem.J., 433, 2010
|
|
