7TO2
 
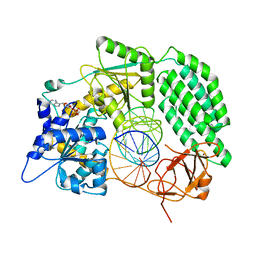 | |
7TO1
 
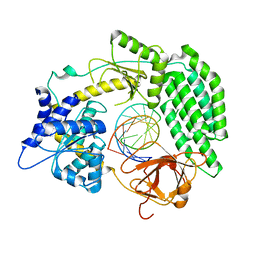 | |
7TO0
 
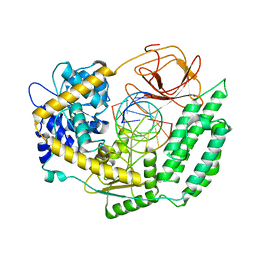 | | Cryo-EM structure of RIG-I in complex with OHdsRNA | | 分子名称: | Antiviral innate immune response receptor RIG-I, OHdsRNA, ZINC ION | | 著者 | Wang, W, Pyle, A.M. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNZ
 
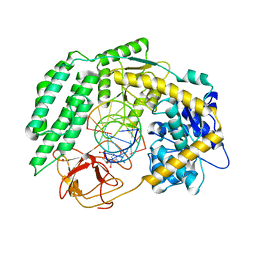 | | Cryo-EM structure of RIG-I in complex with p1dsRNA | | 分子名称: | Antiviral innate immune response receptor RIG-I, ZINC ION, p1dsRNA | | 著者 | Wang, W, Pyle, A.M. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
6A2W
 
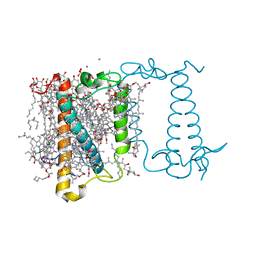 | | Crystal structure of fucoxanthin chlorophyll a/c complex from Phaeodactylum tricornutum | | 分子名称: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | 著者 | Wang, W, Yu, L.J, Kuang, T.Y, Shen, J.R. | | 登録日 | 2018-06-13 | | 公開日 | 2019-02-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for blue-green light harvesting and energy dissipation in diatoms.
Science, 363, 2019
|
|
8K8T
 
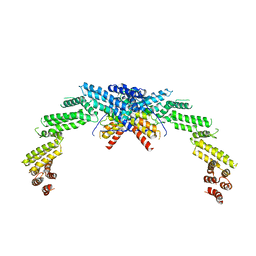 | | Structure of CUL3-RBX1-KLHL22 complex | | 分子名称: | Cullin-3, Kelch-like protein 22 | | 著者 | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | 登録日 | 2023-07-31 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
8K9I
 
 | | Structure of CUL3-RBX1-KLHL22 complex without CUL3 NA motif | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | 著者 | Wang, W, Ling, L, Dai, Z, Zuo, P, Yin, Y. | | 登録日 | 2023-08-01 | | 公開日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | A conserved N-terminal motif of CUL3 contributes to assembly and E3 ligase activity of CRL3 KLHL22.
Nat Commun, 15, 2024
|
|
8G7U
 
 | |
8G7T
 
 | |
8G7V
 
 | |
2I1L
 
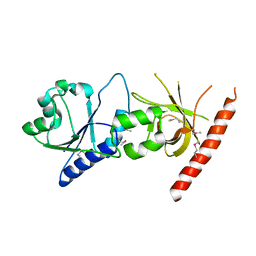 | | Crystal structure of the C2 form of FAD synthetase from Thermotoga maritima | | 分子名称: | Riboflavin kinase/FMN adenylyltransferase | | 著者 | Wang, W, Shin, D.H, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2006-08-14 | | 公開日 | 2006-11-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the C2 form of FAD synthetase from Thermotoga maritima
To be Published
|
|
7D5I
 
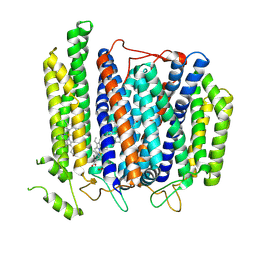 | | Structure of Mycobacterium smegmatis bd complex in the apo-form. | | 分子名称: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome D ubiquinol oxidase subunit 1, HEME B/C, ... | | 著者 | Wang, W, Gong, H, Gao, Y, Zhou, X, Rao, Z. | | 登録日 | 2020-09-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.79 Å) | | 主引用文献 | Cryo-EM structure of mycobacterial cytochrome bd reveals two oxygen access channels.
Nat Commun, 12, 2021
|
|
7BV8
 
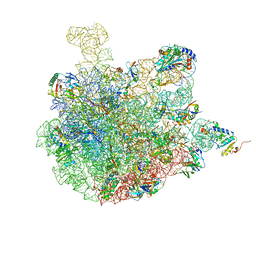 | | Mature 50S ribosomal subunit from RrmJ knock out E.coli strain | | 分子名称: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | 著者 | Wang, W, Li, W.Q, Ge, X.L, Yan, K.G, Mandava, C.S, Sanyal, S, Gao, N. | | 登録日 | 2020-04-09 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Loss of a single methylation in 23S rRNA delays 50S assembly at multiple late stages and impairs translation initiation and elongation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6J0Z
 
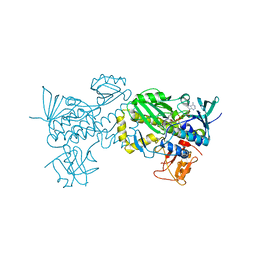 | | Crystal structure of AlpK | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Putative angucycline-like polyketide oxygenase | | 著者 | Wang, W, Liu, Y, Liang, H. | | 登録日 | 2018-12-27 | | 公開日 | 2019-03-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.889 Å) | | 主引用文献 | Crystal structure of AlpK: An essential monooxygenase involved in the biosynthesis of kinamycin
Biochem. Biophys. Res. Commun., 510, 2019
|
|
4FXX
 
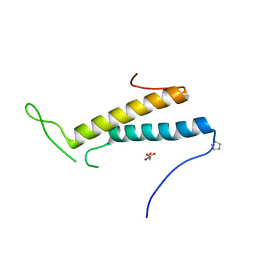 | | Structure of SF1 coiled-coil domain | | 分子名称: | IMIDAZOLE, MALONATE ION, Splicing factor 1 | | 著者 | Gupta, A, Bauer, W.J, Wang, W, Kielkopf, C.L. | | 登録日 | 2012-07-03 | | 公開日 | 2013-01-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4801 Å) | | 主引用文献 | Structure of Phosphorylated SF1 Bound to U2AF(65) in an Essential Splicing Factor Complex.
Structure, 21, 2013
|
|
3SOV
 
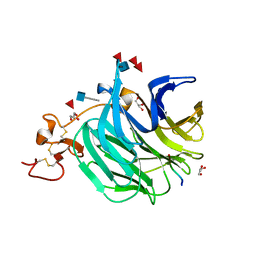 | | The structure of a beta propeller domain in complex with peptide S | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Low-density lipoprotein receptor-related protein 6, ... | | 著者 | Wang, W, Bourhis, E, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | 登録日 | 2011-06-30 | | 公開日 | 2011-09-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
3SOQ
 
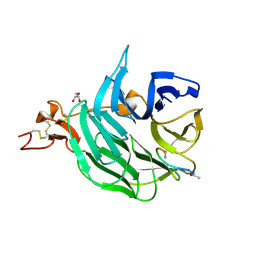 | | The structure of the first YWTD beta propeller domain of LRP6 in complex with a DKK1 peptide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Dickkopf-related protein 1, ... | | 著者 | Wang, W, Bourhis, E, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | 登録日 | 2011-06-30 | | 公開日 | 2011-09-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
3SOB
 
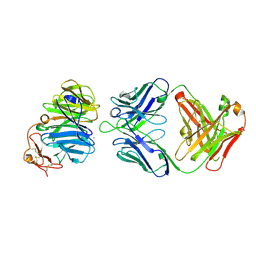 | | The structure of the first YWTD beta propeller domain of LRP6 in complex with a FAB | | 分子名称: | CALCIUM ION, Low-density lipoprotein receptor-related protein 6, antibody heavy chain, ... | | 著者 | Wang, W, Bourhis, E, Tam, C, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | 登録日 | 2011-06-30 | | 公開日 | 2011-09-21 | | 最終更新日 | 2014-05-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
2VG4
 
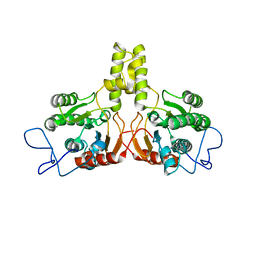 | | Rv2361 native | | 分子名称: | UNDECAPRENYL PYROPHOSPHATE SYNTHETASE | | 著者 | Naismith, J.H, Wang, W, Dong, C. | | 登録日 | 2007-11-08 | | 公開日 | 2007-11-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
2VG1
 
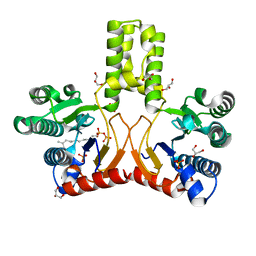 | | Rv1086 E,E-farnesyl diphosphate complex | | 分子名称: | FARNESYL DIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Naismith, J.H, Wang, W, Dong, C. | | 登録日 | 2007-11-07 | | 公開日 | 2007-11-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
4XZC
 
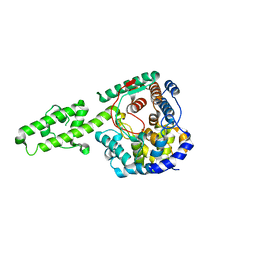 | | The crystal structure of Kupe virus nucleoprotein | | 分子名称: | Nucleoprotein | | 著者 | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | 登録日 | 2015-02-04 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
2VG0
 
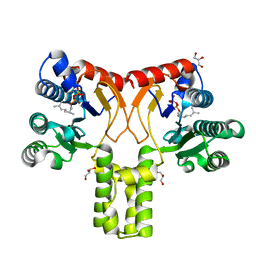 | |
4XZE
 
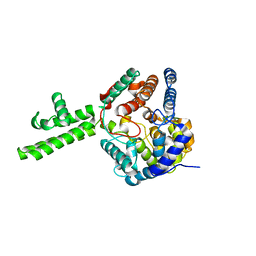 | | The crystal structure of Hazara virus nucleoprotein | | 分子名称: | Nucleoprotein | | 著者 | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | 登録日 | 2015-02-04 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
4XZA
 
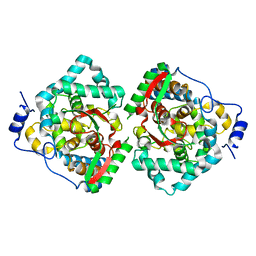 | | The crystal structure of Erve virus nucleoprotein | | 分子名称: | Nucleoprotein | | 著者 | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | 登録日 | 2015-02-04 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
4XZ8
 
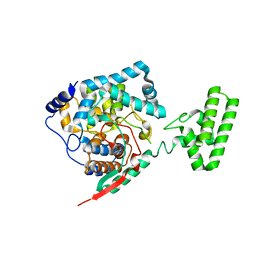 | | The crystal structure of Erve virus nucleoprotein | | 分子名称: | Nucleoprotein | | 著者 | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | 登録日 | 2015-02-04 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
