7E8M
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with mutated RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | 著者 | Wang, X.Q, Zhang, L.Q, Ge, J.W, Wang, R.K, Lan, J. | | 登録日 | 2021-03-02 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species.
Immunity, 54, 2021
|
|
5GT1
 
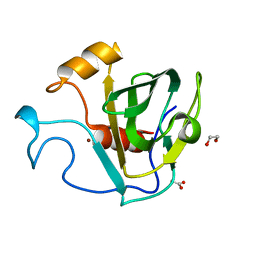 | | crystal structure of cbpa from L. salivarius REN | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Choline binding protein A, ... | | 著者 | Jiang, L, Ren, F. | | 登録日 | 2016-08-18 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Adhesion of Lactobacillus salivarius REN to a Human Intestinal Epithelial Cell Line Requires S-layer Proteins
Sci Rep, 7, 2017
|
|
5HN2
 
 | |
5HNJ
 
 | |
5HNQ
 
 | |
5XP6
 
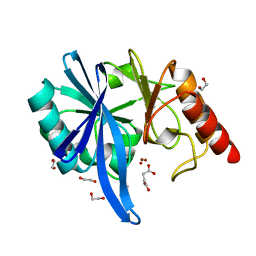 | | native structure of NDM-1 crystallized at pH5.5 | | 分子名称: | 1,2-ETHANEDIOL, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | 著者 | Zhang, H, Ma, G, Lai, J, Sun, H. | | 登録日 | 2017-06-01 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Bismuth antimicrobial drugs serve as broad-spectrum metallo-beta-lactamase inhibitors
Nat Commun, 9, 2018
|
|
5XP9
 
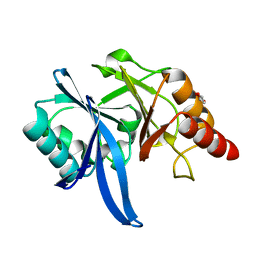 | | Crystal structure of Bismuth bound NDM-1 | | 分子名称: | Bismuth(III) ION, GLYCEROL, Metallo-beta-lactamase type 2 | | 著者 | Zhang, H, Ma, G, Lai, T.P, Sun, H. | | 登録日 | 2017-06-01 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Bismuth antimicrobial drugs serve as broad-spectrum metallo-beta-lactamase inhibitors
Nat Commun, 9, 2018
|
|
2L8E
 
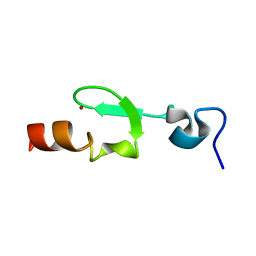 | |
5XVA
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH216 | | 分子名称: | ETHANOL, Serine/threonine-protein kinase PAK 4, [6-chloranyl-4-[(5-methyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl]-[(3R)-3-methylpiperazin-1-yl]methanone | | 著者 | Zhao, F, Li, H. | | 登録日 | 2017-06-27 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.847 Å) | | 主引用文献 | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XVG
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH226 | | 分子名称: | 1,2-ETHANEDIOL, ETHANOL, Serine/threonine-protein kinase PAK 4, ... | | 著者 | Zhao, F, Li, H. | | 登録日 | 2017-06-27 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5X7U
 
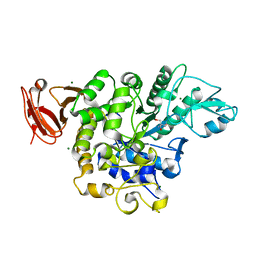 | | Trehalose synthase from Thermobaculum terrenum | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Trehalose synthase | | 著者 | Su, J, Wang, F. | | 登録日 | 2017-02-27 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Structural Characteristics and Function of a New Kind of Thermostable Trehalose Synthase from Thermobaculum terrenum.
J. Agric. Food Chem., 65, 2017
|
|
6LAD
 
 | |
6LAF
 
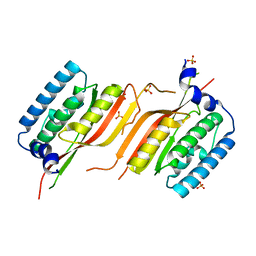 | | Crystal structure of the core domain of Amuc_1100 from Akkermansia muciniphila | | 分子名称: | Amuc_1100, SULFATE ION | | 著者 | Wang, J, Xiang, R, Zhang, M, Wang, M. | | 登録日 | 2019-11-12 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.001 Å) | | 主引用文献 | The variable oligomeric state of Amuc_1100 from Akkermansia muciniphila.
J.Struct.Biol., 212, 2020
|
|
7T62
 
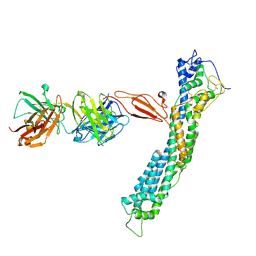 | | GPC2 HEP CT3 complex | | 分子名称: | CT3, Glypican-2 | | 著者 | Zhu, J, Cachau, R, De Val Alda, N, Li, N, Ho, M. | | 登録日 | 2021-12-13 | | 公開日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (21 Å) | | 主引用文献 | CAR T cells targeting tumor-associated exons of glypican 2 regress neuroblastoma in mice.
Cell Rep Med, 2, 2021
|
|
3KJ4
 
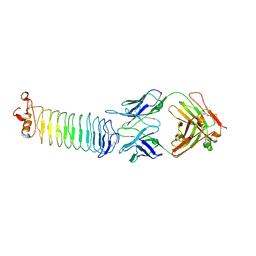 | | Structure of rat Nogo receptor bound to 1D9 antagonist antibody | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab fragment 1D9 heavy chain, ... | | 著者 | Silvian, L.F. | | 登録日 | 2009-11-02 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Resolution of disulfide heterogeneity in Nogo receptor 1 fusion proteins by molecular engineering.
Biotechnol Appl Biochem, 57, 2010
|
|
5WXH
 
 | | Crystal structure of TAF3 PHD finger bound to H3K4me3 | | 分子名称: | Histone H3K4me3, Transcription initiation factor TFIID subunit 3, ZINC ION | | 著者 | Zhao, S, Huang, J, Li, H. | | 登録日 | 2017-01-07 | | 公開日 | 2017-08-16 | | 最終更新日 | 2017-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.297 Å) | | 主引用文献 | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WXG
 
 | | Structure of TAF PHD finger domain binds to H3(1-15)K4ac | | 分子名称: | Histone H3K4ac, MAGNESIUM ION, Transcription initiation factor TFIID subunit 3, ... | | 著者 | Zhao, S, Li, H. | | 登録日 | 2017-01-07 | | 公開日 | 2017-08-16 | | 最終更新日 | 2017-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.703 Å) | | 主引用文献 | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8VB1
 
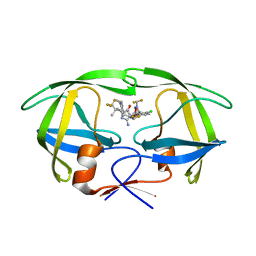 | | Crystal structure of HIV-1 protease with GS-9770 | | 分子名称: | (2S)-2-{(3M)-4-chloro-3-[1-(difluoromethyl)-1H-1,2,4-triazol-5-yl]phenyl}-2-[(2E,4R)-4-[4-(2-cyclopropyl-2H-1,2,3-triazol-4-yl)phenyl]-2-imino-5-oxo-4-(3,3,3-trifluoro-2,2-dimethylpropyl)imidazolidin-1-yl]ethyl [1-(difluoromethyl)cyclopropyl]carbamate, HIV-1 protease | | 著者 | Lansdon, E.B. | | 登録日 | 2023-12-11 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Preclinical characterization of a non-peptidomimetic HIV protease inhibitor with improved metabolic stability.
Antimicrob.Agents Chemother., 68, 2024
|
|
4XGY
 
 | | GFP based antibody (fluorobody) | | 分子名称: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Green fluorescent protein, mAb LCDR3 | | 著者 | Shi, N, Chen, Y.G, Wang, S.H. | | 登録日 | 2015-01-03 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.494 Å) | | 主引用文献 | The structure of a GFP-based antibody (fluorobody) to TLH, a toxin from Vibrio parahaemolyticus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6LHE
 
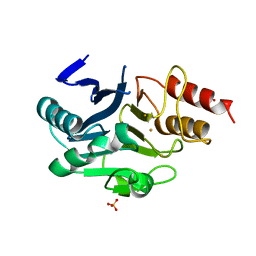 | | Crystal Structure of Gold-bound NDM-1 | | 分子名称: | GOLD ION, Metallo-beta-lactamase type 2, SULFATE ION | | 著者 | Wang, H, Sun, H, Wang, M. | | 登録日 | 2019-12-07 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.206 Å) | | 主引用文献 | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
6LI6
 
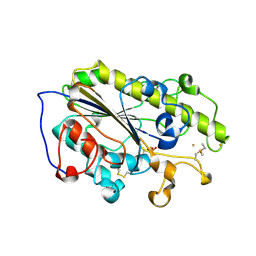 | |
6LI5
 
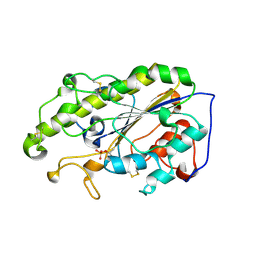 | | Crystal structure of apo-MCR-1-S | | 分子名称: | Probable phosphatidylethanolamine transferase Mcr-1 | | 著者 | Zhang, Q, Wang, M, Sun, H. | | 登録日 | 2019-12-10 | | 公開日 | 2020-09-16 | | 最終更新日 | 2020-10-28 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
6LI4
 
 | | Crystal structure of MCR-1-S | | 分子名称: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | 著者 | Zhang, Q, Wang, M, Sun, H. | | 登録日 | 2019-12-10 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
5XM8
 
 | |
8X84
 
 | |
