6OA1
 
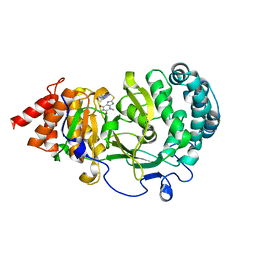 | | Structure of human PARG complexed with JA2120 | | 分子名称: | 1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]sulfanyl}-3,7-dihydro-1H-purine-2,6-dione, Poly(ADP-ribose) glycohydrolase | | 著者 | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-12-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
6ASI
 
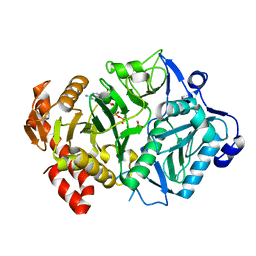 | | E. coli phosphoenolpyruvate carboxykinase G209S mutant bound to methanesulfonate | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | 著者 | Tang, H.Y.H, Shin, D.S, Tainer, J.A. | | 登録日 | 2017-08-24 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.789 Å) | | 主引用文献 | Structural Control of Nonnative Ligand Binding in Engineered Mutants of Phosphoenolpyruvate Carboxykinase.
Biochemistry, 57, 2018
|
|
6AT2
 
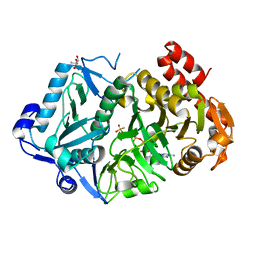 | | E. coli phosphoenolpyruvate carboxykinase G209N mutant bound to thiosulfate | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Tang, H.Y.H, Shin, D.S, Tainer, J.A. | | 登録日 | 2017-08-27 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.444 Å) | | 主引用文献 | Structural Control of Nonnative Ligand Binding in Engineered Mutants of Phosphoenolpyruvate Carboxykinase.
Biochemistry, 57, 2018
|
|
6ASC
 
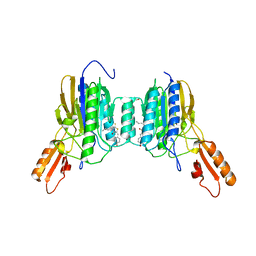 | | Mre11 dimer in complex with Endonuclease inhibitor PFM04 | | 分子名称: | (5E)-3-butyl-5-[(4-hydroxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, 1,2-ETHANEDIOL, MANGANESE (II) ION, ... | | 著者 | Moiani, D, Arvai, A.S, Tainer, J.A. | | 登録日 | 2017-08-24 | | 公開日 | 2018-03-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Targeting Allostery with Avatars to Design Inhibitors Assessed by Cell Activity: Dissecting MRE11 Endo- and Exonuclease Activities.
Meth. Enzymol., 601, 2018
|
|
6PBK
 
 | |
6AX6
 
 | | The crystal structure of a lysyl hydroxylase from Acanthamoeba polyphaga mimivirus | | 分子名称: | FE (II) ION, IODIDE ION, Procollagen lysyl hydroxylase and glycosyltransferase | | 著者 | Guo, H, Tsai, C, Miller, M.D, Alvarado, S, Tainer, J.A, Phillips Jr, G.N, Kurie, J.M. | | 登録日 | 2017-09-06 | | 公開日 | 2018-02-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.241 Å) | | 主引用文献 | Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding.
Nat Commun, 9, 2018
|
|
6ASN
 
 | |
6ASM
 
 | | E. coli phosphoenolpyruvate carboxykinase G209S K212C mutant bound to thiosulfate | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Tang, H.Y.H, Shin, D.S, Tainer, J.A. | | 登録日 | 2017-08-25 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural Control of Nonnative Ligand Binding in Engineered Mutants of Phosphoenolpyruvate Carboxykinase.
Biochemistry, 57, 2018
|
|
6AT4
 
 | |
6AT3
 
 | |
4HDV
 
 | |
2AV8
 
 | |
6AX7
 
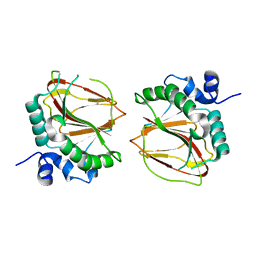 | | The crystal structure of a lysyl hydroxylase from Acanthamoeba polyphaga mimivirus | | 分子名称: | FE (II) ION, Procollagen lysyl hydroxylase and glycosyltransferase | | 著者 | Guo, H, Tsai, C, Miller, M.D, Alvarado, S, Tainer, J.A, Phillips Jr, G.N, Kurie, J.M. | | 登録日 | 2017-09-06 | | 公開日 | 2018-02-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding.
Nat Commun, 9, 2018
|
|
2FC0
 
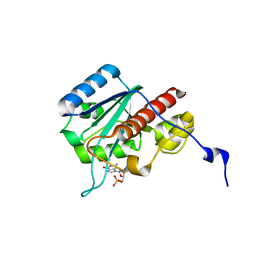 | | WRN exonuclease, Mn dGMP complex | | 分子名称: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Werner syndrome helicase | | 著者 | Perry, J.J, Tainer, J.A. | | 登録日 | 2005-12-10 | | 公開日 | 2006-04-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FWQ
 
 | |
3Q8L
 
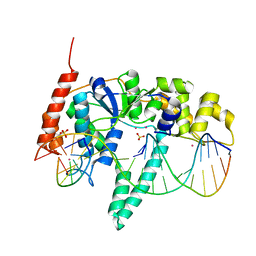 | | Crystal Structure of Human Flap Endonuclease FEN1 (WT) in complex with substrate 5'-flap DNA, SM3+, and K+ | | 分子名称: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | 著者 | Tsutakawa, S.E, Classen, S, Chapados, B.R, Arvai, A, Finger, D.L, Guenther, G, Tomlinson, C.G, Thompson, P, Sarker, A.H, Shen, B, Cooper, P.K, Grasby, J.A, Tainer, J.A. | | 登録日 | 2011-01-06 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.319 Å) | | 主引用文献 | Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a Unified Understanding of the FEN1 Superfamily.
Cell(Cambridge,Mass.), 145, 2011
|
|
1YHI
 
 | | Uncyclized precursor structure of S65A Y66S R96A GFP variant | | 分子名称: | Green Fluorescent Protein | | 著者 | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2005-01-08 | | 公開日 | 2005-02-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Understanding GFP Chromophore Biosynthesis: Controlling Backbone Cyclization and Modifying Post-translational Chemistry(,).
Biochemistry, 44, 2005
|
|
1YJ2
 
 | | Cyclized, non-dehydrated post-translational product for S65A Y66S H148G GFP variant | | 分子名称: | 1,2-ETHANEDIOL, Green Fluorescent Protein, MAGNESIUM ION | | 著者 | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2005-01-13 | | 公開日 | 2005-02-15 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Understanding GFP Chromophore Biosynthesis: Controlling Backbone Cyclization and Modifying Post-translational Chemistry.
Biochemistry, 44, 2005
|
|
1YJF
 
 | | Cyclized post-translational product for S65A Y66S (GFPhal) green fluorescent protein variant | | 分子名称: | Green Fluorescent Protein, MAGNESIUM ION | | 著者 | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2005-01-14 | | 公開日 | 2005-02-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Understanding GFP Chromophore Biosynthesis: Controlling Backbone Cyclization and Modifying Post-translational Chemistry.
Biochemistry, 44, 2005
|
|
3Q8M
 
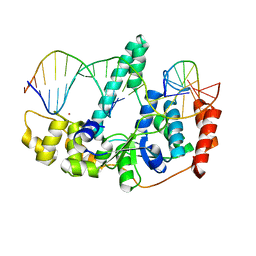 | | Crystal Structure of Human Flap Endonuclease FEN1 (D181A) in complex with substrate 5'-flap DNA and K+ | | 分子名称: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | 著者 | Tsutakawa, S.E, Classen, S, Chapados, B.R, Arvai, A, Finger, D.L, Guenther, G, Tomlinson, C.G, Thompson, P, Sarker, A.H, Shen, B, Cooper, P.K, Grasby, J.A, Tainer, J.A. | | 登録日 | 2011-01-06 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a Unified Understanding of the FEN1 Superfamily.
Cell(Cambridge,Mass.), 145, 2011
|
|
1YHG
 
 | |
3Q8K
 
 | | Crystal Structure of Human Flap Endonuclease FEN1 (WT) in complex with product 5'-flap DNA, SM3+, and K+ | | 分子名称: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(P*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | 著者 | Tsutakawa, S.E, Classen, S, Chapados, B.R, Arvai, A, Finger, D.L, Guenther, G, Tomlinson, C.G, Thompson, P, Sarker, A.H, Shen, B, Cooper, P.K, Grasby, J.A, Tainer, J.A. | | 登録日 | 2011-01-06 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2001 Å) | | 主引用文献 | Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a Unified Understanding of the FEN1 Superfamily.
Cell(Cambridge,Mass.), 145, 2011
|
|
1YAI
 
 | | X-RAY STRUCTURE OF A BACTERIAL COPPER,ZINC SUPEROXIDE DISMUTASE | | 分子名称: | COPPER (II) ION, COPPER, ZINC SUPEROXIDE DISMUTASE, ... | | 著者 | Bourne, Y, Redford, S.M, Lo, T.P, Tainer, J.A, Getzoff, E.D. | | 登録日 | 1996-02-03 | | 公開日 | 1997-08-20 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Novel dimeric interface and electrostatic recognition in bacterial Cu,Zn superoxide dismutase.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1YHH
 
 | |
2UGI
 
 | | PROTEIN MIMICRY OF DNA FROM CRYSTAL STRUCTURES OF THE URACIL GLYCOSYLASE INHIBITOR PROTEIN AND ITS COMPLEX WITH ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE | | 分子名称: | IMIDAZOLE, URACIL-DNA GLYCOSYLASE INHIBITOR | | 著者 | Putnam, C.D, Arvai, A.S, Mol, C.D, Tainer, J.A. | | 登録日 | 1998-11-06 | | 公開日 | 1999-03-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
