2MQ0
 
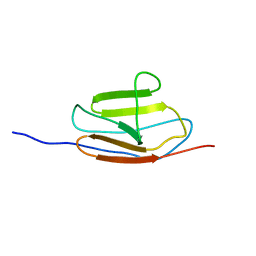 | | NMR structure of the c3 domain of human cardiac myosin binding protein-c | | 分子名称: | Myosin-binding protein C, cardiac-type | | 著者 | Zhang, X, De, S, Mcintosh, L.P, Paetzel, M. | | 登録日 | 2014-06-10 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Characterization of the C3 Domain of Cardiac Myosin Binding Protein C and Its Hypertrophic Cardiomyopathy-Related R502W Mutant.
Biochemistry, 53, 2014
|
|
2MQ3
 
 | | NMR structure of the c3 domain of human cardiac myosin binding protein-c with a hypertrophic cardiomyopathy-related mutation R502W. | | 分子名称: | Myosin-binding protein C, cardiac-type | | 著者 | Zhang, X, De, S, Mcintosh, L.P, Paetzel, M. | | 登録日 | 2014-06-12 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Characterization of the C3 Domain of Cardiac Myosin Binding Protein C and Its Hypertrophic Cardiomyopathy-Related R502W Mutant.
Biochemistry, 53, 2014
|
|
7D5K
 
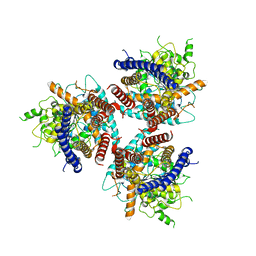 | | CryoEM structure of cotton cellulose synthase isoform 7 | | 分子名称: | Cellulose synthase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Guan, Z.Y, Xue, Y, Yin, P, Zhang, X.L. | | 登録日 | 2020-09-26 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural insights into homotrimeric assembly of cellulose synthase CesA7 from Gossypium hirsutum.
Plant Biotechnol J, 19, 2021
|
|
5ZCX
 
 | | Structure of T20/N39 | | 分子名称: | Envelope glycoprotein, Envelope glycoprotein gp160 | | 著者 | Zhang, X.J, Ding, X.H. | | 登録日 | 2018-02-21 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.299 Å) | | 主引用文献 | Structural and functional characterization of HIV-1 cell fusion inhibitor T20.
AIDS, 33, 2019
|
|
4GJE
 
 | |
4GJF
 
 | |
4GJG
 
 | |
1DYC
 
 | |
1DYD
 
 | |
1DYB
 
 | |
1DYE
 
 | |
1DYA
 
 | |
3RO4
 
 | |
8V57
 
 | |
8V58
 
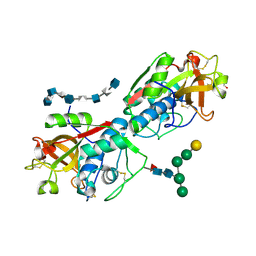 | | Complex of murine cathepsin K with bound heparan sulfate 12mer | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ... | | 著者 | Pedersen, L.C, Xu, D, Krahn, J.M. | | 登録日 | 2023-11-30 | | 公開日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Heparan sulfate selectively inhibits the collagenase activity of cathepsin K.
Matrix Biol., 129, 2024
|
|
4ISI
 
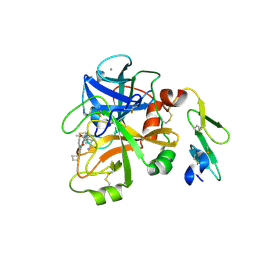 | | Structure of FACTOR VIIA in complex with the inhibitor (6S)-N-(4-CARBAMIMIDOYLBENZYL)-1-CHLORO-3-(CYCLOBUTYLAMINO)-8,8-DIETHYL-4-OXO-4,6,7,8-TETRAHYDROPYRROLO[1,2-A]PYRAZINE-6-CARBOXAMIDE | | 分子名称: | (6S)-N-(4-carbamimidoylbenzyl)-1-chloro-3-(cyclobutylamino)-8,8-diethyl-4-oxo-4,6,7,8-tetrahydropyrrolo[1,2-a]pyrazine-6-carboxamide, CALCIUM ION, Factor VII heavy chain, ... | | 著者 | Wei, A. | | 登録日 | 2013-01-16 | | 公開日 | 2013-02-27 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Design and synthesis of bicyclic pyrazinone and pyrimidinone amides as potent TF-FVIIa inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7EGN
 
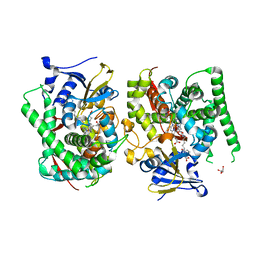 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | 分子名称: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | 著者 | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | 登録日 | 2021-03-24 | | 公開日 | 2021-08-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | H-Bonding Networks Dictate the Molecular Mechanism of H2O2 Activation by P450
Acs Catalysis, 11, 2021
|
|
3UDC
 
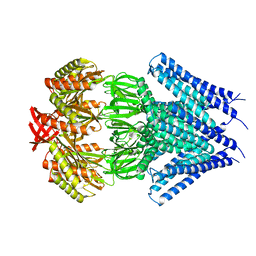 | | Crystal structure of a membrane protein | | 分子名称: | Small-conductance mechanosensitive channel, C-terminal peptide from Small-conductance mechanosensitive channel | | 著者 | Li, W, Ge, J, Yang, M. | | 登録日 | 2011-10-28 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.355 Å) | | 主引用文献 | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6IMR
 
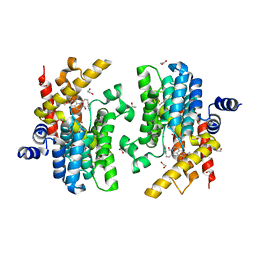 | | Crystal structure of PDE4D complexed with a novel inhibitor | | 分子名称: | (1S)-1-[3-(1H-indol-3-yl)propyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | 著者 | Zhang, X.L, Su, H.X, Xu, Y.C. | | 登録日 | 2018-10-23 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.503 Å) | | 主引用文献 | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IM6
 
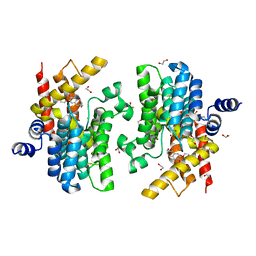 | | Crystal structure of PDE4D complexed with a novel inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 7-ethoxy-6-methoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | 著者 | Zhang, X.L, Su, H.X, Xu, Y.C. | | 登録日 | 2018-10-22 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.702 Å) | | 主引用文献 | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMT
 
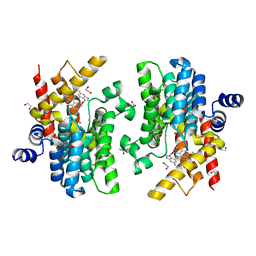 | | Crystal structure of PDE4D complexed with a novel inhibitor | | 分子名称: | (1S)-1-[2-(6-fluoro-1H-indol-3-yl)ethyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | 著者 | Zhang, X.L, Su, H.X, Xu, Y.C. | | 登録日 | 2018-10-23 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.483 Å) | | 主引用文献 | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMO
 
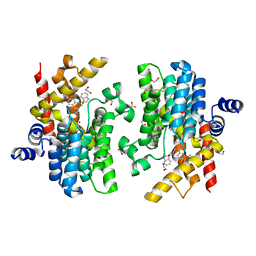 | | Crystal structure of PDE4D complexed with a novel inhibitor | | 分子名称: | (1S)-1-[(1H-indol-3-yl)methyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | 著者 | Zhang, X.L, Su, H.X, Xu, Y.C. | | 登録日 | 2018-10-23 | | 公開日 | 2019-10-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6INM
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | 分子名称: | (1S)-1-[2-(1H-indol-3-yl)ethyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | 著者 | Zhang, X.L, Su, H.X, Xu, Y.C. | | 登録日 | 2018-10-26 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IND
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | 分子名称: | (1S)-6,7-dimethoxy-1-[2-(6-methyl-1H-indol-3-yl)ethyl]-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | 著者 | Zhang, X.L, Su, H.X, Xu, Y.C. | | 登録日 | 2018-10-24 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.872 Å) | | 主引用文献 | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
7VT0
 
 | | Dimer structure of SORLA | | 分子名称: | Sortilin-related receptor | | 著者 | Xi, Z, Cang, W, Chuang, L. | | 登録日 | 2021-10-27 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures reveal distinct apo conformations of sortilin-related receptor SORLA.
Biochem.Biophys.Res.Commun., 600, 2022
|
|
