7UUG
 
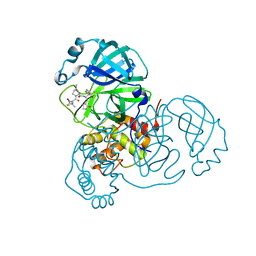 | | SARS-CoV-2 Main Protease S144A (Mpro S144A) in Complex with ML1006a | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | 著者 | Westberg, M, Fernandez, D, Lin, M.Z. | | 登録日 | 2022-04-28 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
7UUP
 
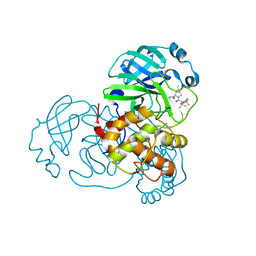 | | SARS-CoV-2 Main Protease S144A (Mpro S144A) in Complex with Nirmatrelvir (PF-07321332) | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | 著者 | Westberg, M, Fernandez, D, Lin, M.Z. | | 登録日 | 2022-04-28 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
7OVU
 
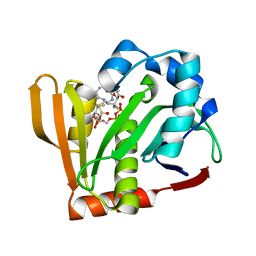 | |
7OVV
 
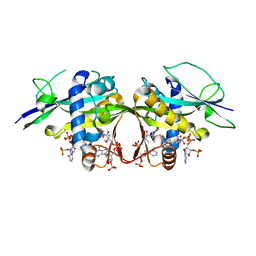 | | Crystal structure of the Arabidopsis thaliana thialysine acetyltransferase AtNATA2 | | 分子名称: | Probable acetyltransferase NATA1-like, [[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyldisulfanyl]ethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | 著者 | Layer, D, Kopp, J, Sinning, I. | | 登録日 | 2021-06-15 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural insights into the Arabidopsis thaliana thialysine acetyltransferase AtNATA2
To Be Published
|
|
7V6Z
 
 | | Cryo-EM structure of Patched1 (V1084A mutant) in lipid nanodisc, 3.64 angstrom (reprocessed with the dataset of 7dzp) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Luo, Y, Zhao, Y, Qu, Q, Li, D. | | 登録日 | 2021-08-20 | | 公開日 | 2021-09-22 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Cryo-EM study of patched in lipid nanodisc suggests a structural basis for its clustering in caveolae.
Structure, 29, 2021
|
|
5ZYI
 
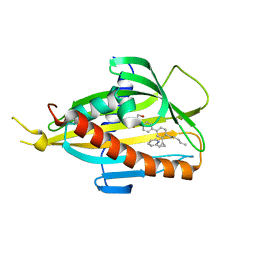 | | Crystal structure of CERT START domain in complex with compound E16 | | 分子名称: | 2-[4-[4-pentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | 著者 | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | 登録日 | 2018-05-25 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
7P5L
 
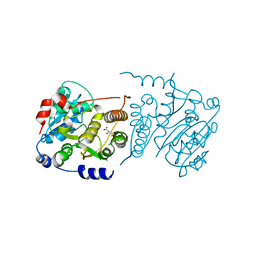 | |
7V6Y
 
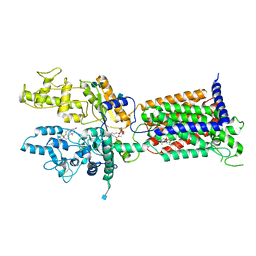 | | Cryo-EM structure of Patched in lipid nanodisc - the wildtype, 3.5 angstrom (re-processed with dataset of 7dzq) | | 分子名称: | (2S)-2-azanyl-3-[[(2S)-3-butanoyloxy-2-dec-9-enoyloxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Luo, Y, Zhao, Y, Qu, Q, Li, D. | | 登録日 | 2021-08-20 | | 公開日 | 2021-09-22 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM study of patched in lipid nanodisc suggests a structural basis for its clustering in caveolae.
Structure, 29, 2021
|
|
6A9N
 
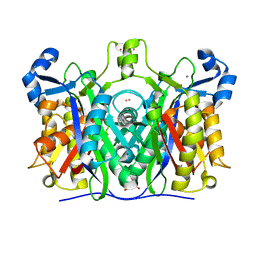 | |
7P8G
 
 | | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 5.5 - apo form | | 分子名称: | CHLORIDE ION, Glucosyl-3-phosphoglycerate synthase, MALONATE ION, ... | | 著者 | Silva, A, Nunes-Costa, D, Barbosa Pereira, P.J, Macedo-Ribeiro, S. | | 登録日 | 2021-07-21 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Crystal structure of Mycobacterium hassiacum glucosyl-3-phosphoglycerate synthase at pH 5.5 - apo form
To Be Published
|
|
7P2K
 
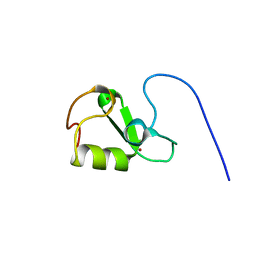 | | Solution NMR Structure of Arginine to Cysteine mutant of Arkadia RING domain. | | 分子名称: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | 著者 | Raptis, V, Marousis, K.D, Birkou, M, Bentrop, D, Episkopou, V, Spyroulias, G.A. | | 登録日 | 2021-07-06 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Impact of a Single Nucleotide Polymorphism on the 3D Protein Structure and Ubiquitination Activity of E3 Ubiquitin Ligase Arkadia.
Front Mol Biosci, 9, 2022
|
|
5ZU2
 
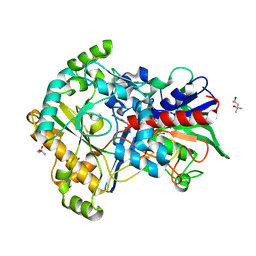 | | Effect of mutation (R554A) on FAD modification in Aspergillus oryzae RIB40formate oxidase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Mikami, B, Uchida, H, Doubayashi, D. | | 登録日 | 2018-05-06 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.441 Å) | | 主引用文献 | The microenvironment surrounding FAD mediates its conversion to 8-formyl-FAD in Aspergillus oryzae RIB40 formate oxidase.
J.Biochem., 166, 2019
|
|
5OVF
 
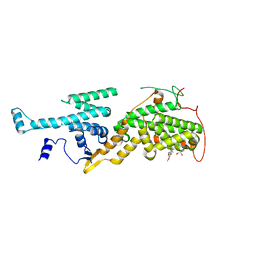 | | Ras guanine nucleotide exchange factor SOS1 (Rem-cdc25) in complex with small molecule inhibitor compound 17 | | 分子名称: | 1,2-ETHANEDIOL, 6,7-dimethoxy-2-methyl-~{N}-[(1~{R})-1-[3-(1~{H}-pyrazol-4-yl)phenyl]ethyl]quinazolin-4-amine, Son of sevenless homolog 1 | | 著者 | Hillig, R.C, Sautier, B, Schroeder, J, Moosmayer, D, Hilpmann, A, Stegmann, C.M, Briem, H, Boemer, U, Weiske, J, Badock, V, Petersen, K, Kahmann, J, Wegener, D, Bohnke, N, Eis, K, Graham, K, Wortmann, L, von Nussbaum, F, Bader, B. | | 登録日 | 2017-08-28 | | 公開日 | 2019-02-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZWY
 
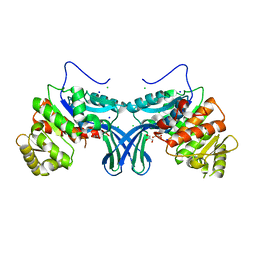 | | Ribokinase from Leishmania donovani | | 分子名称: | CHLORIDE ION, GLYCEROL, Ribokinase, ... | | 著者 | Gatreddi, S, Vasudevan, D, Qureshi, I.A. | | 登録日 | 2018-05-17 | | 公開日 | 2019-05-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Unraveling structural insights of ribokinase from Leishmania donovani.
Int.J.Biol.Macromol., 136, 2019
|
|
5RL1
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-27 (Mpro-x3113) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-tert-butylphenyl)-N-[(1R)-2-[(3-methoxypropyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-08-05 | | 公開日 | 2020-12-02 | | 最終更新日 | 2021-07-07 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
7VKO
 
 | | Crystal structure of TrkA kinase with repotrectinib | | 分子名称: | Repotrectinib, SULFATE ION, Tyrosine-protein kinase receptor | | 著者 | Murray, B.W, Rogers, E, Zhai, D, Deng, W, Chen, X, Sprengeler, P.A, Zhang, X, Graber, A, Reich, S.H, Stopatschinskaja, S, Solomon, B, Besse, B, Drilon, A. | | 登録日 | 2021-09-30 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Molecular Characteristics of Repotrectinib That Enable Potent Inhibition of TRK Fusion Proteins and Resistant Mutations.
Mol.Cancer Ther., 20, 2021
|
|
7VKM
 
 | | Crystal structure of TrkA (G595R) kinase domain | | 分子名称: | Tyrosine-protein kinase receptor | | 著者 | Murray, B.W, Rogers, E, Zhai, D, Deng, W, Chen, X, Sprengeler, P.A, Zhang, X, Graber, A, Reich, S.H, Stopatschinskaja, S, Solomon, B, Besse, B, Drilon, A. | | 登録日 | 2021-09-30 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Molecular Characteristics of Repotrectinib That Enable Potent Inhibition of TRK Fusion Proteins and Resistant Mutations.
Mol.Cancer Ther., 20, 2021
|
|
7VKN
 
 | | Crystal structure of TrkA (G595R) kinase with repotrectinib | | 分子名称: | Repotrectinib, SULFATE ION, Tyrosine-protein kinase receptor | | 著者 | Murray, B.W, Rogers, E, Zhai, D, Deng, W, Chen, X, Sprengeler, P.A, Zhang, X, Graber, A, Reich, S.H, Stopatschinskaja, S, Solomon, B, Besse, B, Drilon, A. | | 登録日 | 2021-09-30 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular Characteristics of Repotrectinib That Enable Potent Inhibition of TRK Fusion Proteins and Resistant Mutations.
Mol.Cancer Ther., 20, 2021
|
|
5ZYJ
 
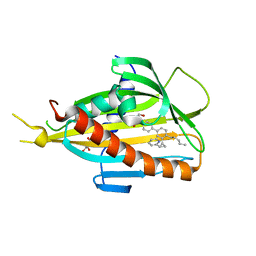 | | Crystal structure of CERT START domain in complex with compound E16A | | 分子名称: | 2-[4-[4-pentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, GLYCEROL, LIPID-TRANSFER PROTEIN CERT | | 著者 | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | 登録日 | 2018-05-25 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZZ2
 
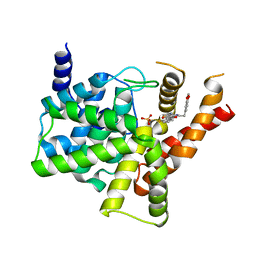 | | Crystal structure of PDE5 in complex with inhibitor LW1634 | | 分子名称: | 3-[(2H-1,3-benzodioxol-5-yl)methyl]-8-fluoro-1-(1,3-thiazol-2-yl)[1]benzopyrano[2,3-c]pyrrol-9(2H)-one, MAGNESIUM ION, SULFATE ION, ... | | 著者 | Wu, D, Huang, Y.D, Huang, Y.Y, Luo, H.B. | | 登録日 | 2018-05-29 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Optimization of Chromeno[2,3- c]pyrrol-9(2 H)-ones as Highly Potent, Selective, and Orally Bioavailable PDE5 Inhibitors: Structure-Activity Relationship, X-ray Crystal Structure, and Pharmacodynamic Effect on Pulmonary Arterial Hypertension.
J. Med. Chem., 61, 2018
|
|
6A8C
 
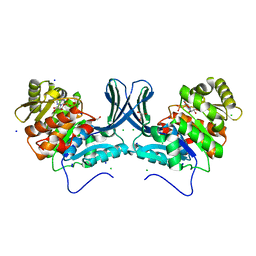 | | Ribokinase from Leishmania donovani with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Gatreddi, S, Vasudevan, D, Qureshi, I.A. | | 登録日 | 2018-07-06 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Unraveling structural insights of ribokinase from Leishmania donovani.
Int.J.Biol.Macromol., 136, 2019
|
|
5OX8
 
 | | Structure of Enhanced Cyan Fluorescent Protein at pH 5.0 | | 分子名称: | CHLORIDE ION, Green fluorescent protein | | 著者 | Gotthard, G, von Stetten, D, Clavel, D, Noirclerc-Savoye, M, Royant, A. | | 登録日 | 2017-09-06 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Chromophore Isomer Stabilization Is Critical to the Efficient Fluorescence of Cyan Fluorescent Proteins.
Biochemistry, 56, 2017
|
|
7Q4M
 
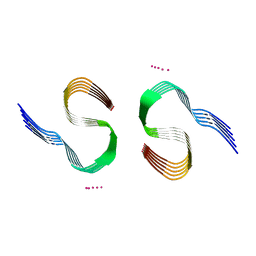 | | Type II beta-amyloid 42 Filaments from Human Brain | | 分子名称: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | 著者 | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | 登録日 | 2021-11-01 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
6A1F
 
 | | Crystal structure of human DYRK1A in complex with compound 14 | | 分子名称: | 8-methoxy-5,5-dimethyl-5,6-dihydrobenzo[h]quinazolin-4-amine, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | 著者 | Baba, D, Hanzawa, H. | | 登録日 | 2018-06-07 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Discovery of DS42450411 as a potent orally active hepcidin production inhibitor: Design and optimization of novel 4-aminopyrimidine derivatives.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
7Q4B
 
 | | Type I beta-amyloid 42 Filaments from Human Brain | | 分子名称: | Amyloid-beta precursor protein, UNKNOWN ATOM OR ION | | 著者 | Yang, Y, Arseni, D, Zhang, W, Huang, M, Lovestam, S.K.A, Schweighauser, M, Kotecha, A, Murzin, A.G, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Garringer, H.J, Gelpi, E, Newell, K.L, Kovacs, G.G, Vidal, R, Ghetti, B, Falcon, B, Scheres, S.H.W, Goedert, M. | | 登録日 | 2021-10-30 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structures of amyloid-beta 42 filaments from human brains.
Science, 375, 2022
|
|
