6V6W
 
 | |
7TO4
 
 | | Structural and functional impact by SARS-CoV-2 Omicron spike mutations | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | 登録日 | 2022-01-22 | | 公開日 | 2022-02-16 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural and functional impact by SARS-CoV-2 Omicron spike mutations.
Cell Rep, 39, 2022
|
|
7TNW
 
 | | Structural and functional impact by SARS-CoV-2 Omicron spike mutations | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | 登録日 | 2022-01-21 | | 公開日 | 2022-02-16 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural and functional impact by SARS-CoV-2 Omicron spike mutations.
Cell Rep, 39, 2022
|
|
5KCC
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with Oxabicyclic Heptene Sulfonamide (OBHS-N) | | 分子名称: | (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-N-phenyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2016-06-06 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.386 Å) | | 主引用文献 | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
6VQV
 
 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF9 | | 分子名称: | AcrF9, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | 著者 | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | 登録日 | 2020-02-06 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VQX
 
 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF6 | | 分子名称: | AcrF6, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | 著者 | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | 登録日 | 2020-02-06 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7TVA
 
 | | Stat5a Core in complex with AK2292 | | 分子名称: | DI(HYDROXYETHYL)ETHER, MALONATE ION, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N-(5-{2-[(3R)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}pent-4-yn-1-yl)-N-methyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, ... | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2022-02-04 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.835 Å) | | 主引用文献 | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
7TVB
 
 | | Stat5A Core in Complex with AK305 | | 分子名称: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N,N-dimethyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, Signal transducer and activator of transcription 5A | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2022-02-04 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.653 Å) | | 主引用文献 | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
7CJF
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | 著者 | Guo, Y, Li, X, Zhang, G, Fu, D, Schweizer, L, Zhang, H, Rao, Z. | | 登録日 | 2020-07-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.108 Å) | | 主引用文献 | A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Nat Commun, 12, 2021
|
|
8GAS
 
 | | vFP48.02 Fab in complex with BG505 DS-SOSIP Env trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Gorman, J, Kwong, P.D. | | 登録日 | 2023-02-23 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (4.04 Å) | | 主引用文献 | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8G85
 
 | | vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Gorman, J, Kwong, P.D. | | 登録日 | 2023-02-17 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (3.99 Å) | | 主引用文献 | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
6G9P
 
 | |
5EM7
 
 | |
6VQW
 
 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF8 | | 分子名称: | AcrF8, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | 著者 | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | 登録日 | 2020-02-06 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5HCY
 
 | | EGFR kinase domain mutant "TMLR" with 3-carboxamide azaindole compound 13 | | 分子名称: | 6-[[2-(1-cyclopropylsulfonylpyrazol-4-yl)pyrimidin-4-yl]amino]-~{N}-(oxan-4-yl)-1-propan-2-yl-pyrrolo[3,2-c]pyridine-3-carboxamide, Epidermal growth factor receptor, SULFATE ION | | 著者 | Eigenbrot, C, Yu, C. | | 登録日 | 2016-01-04 | | 公開日 | 2016-09-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Discovery of a Noncovalent, Mutant-Selective Epidermal Growth Factor Receptor Inhibitor.
J.Med.Chem., 59, 2016
|
|
7UNJ
 
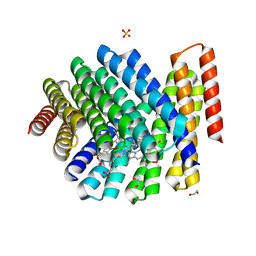 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester matching geometry of purple bacterial special pair, SP1-ZnPPaM | | 分子名称: | 1,2-ETHANEDIOL, SP1-ZnPPaM designed chlorophyll dimer protein, SULFATE ION, ... | | 著者 | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | 登録日 | 2022-04-11 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
7UNH
 
 | |
7UNI
 
 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester, SP2-ZnPPaM | | 分子名称: | 1,2-ETHANEDIOL, PHOSPHATE ION, SP2-ZnPPaM designed chlorophyll dimer protein, ... | | 著者 | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | 登録日 | 2022-04-11 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
5SQ5
 
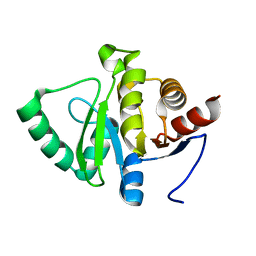 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894407 - (R,S) and (S,S) isomers | | 分子名称: | (1R,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQA
 
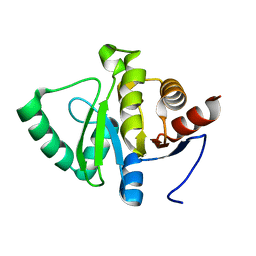 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894395 - (R,R) and (S,S) isomers | | 分子名称: | (1R,2R)-4-hydroxy-1-[(3-oxo-3,4-dihydro-2H-1,4-benzoxazine-7-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-[(3-oxo-3,4-dihydro-2H-1,4-benzoxazine-7-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQ9
 
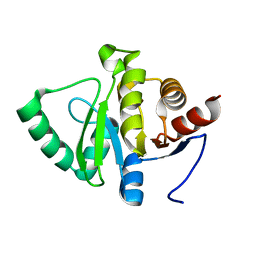 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894420 - (R,R) and (S,S) isomers | | 分子名称: | (1R,2R)-4-hydroxy-1-{4-[(propan-2-yl)carbamamido]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-{4-[(propan-2-yl)carbamamido]benzamido}-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQ7
 
 | |
5SQD
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894382 - (R,R) and (S,S) isomers | | 分子名称: | (1R,2R)-4-hydroxy-1-{[2-(hydroxymethyl)-1H-benzimidazole-5-carbonyl]amino}-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-{[2-(hydroxymethyl)-1H-benzimidazole-5-carbonyl]amino}-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SQH
 
 | |
5SQG
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894430 - (R,R) and (S,S) isomers | | 分子名称: | (1R,2R)-1-{[6-(cyclopropylcarbamamido)pyridine-3-carbonyl]amino}-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-1-{[6-(cyclopropylcarbamamido)pyridine-3-carbonyl]amino}-4-hydroxy-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
