3QJU
 
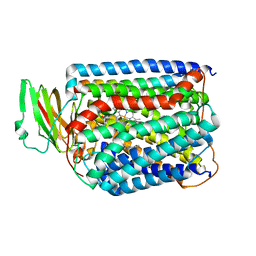 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | 分子名称: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | 著者 | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | 登録日 | 2011-01-30 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
6KM7
 
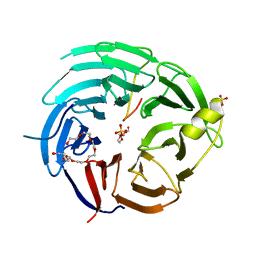 | | The structural basis for the internal interaction in RBBP5 | | 分子名称: | HEXAETHYLENE GLYCOL, NONAETHYLENE GLYCOL, Retinoblastoma-binding protein 5, ... | | 著者 | Han, J, Li, T, Chen, Y. | | 登録日 | 2019-07-31 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | The internal interaction in RBBP5 regulates assembly and activity of MLL1 methyltransferase complex.
Nucleic Acids Res., 47, 2019
|
|
3QJT
 
 | | The structure of and photolytic induced changes of carbon monoxide binding to the cytochrome ba3-oxidase from Thermus thermophilus | | 分子名称: | CARBON MONOXIDE, COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, ... | | 著者 | Liu, B, Zhang, Y, Sage, J.T, Doukov, T, Chen, Y, Stout, C.D, Fee, J.A. | | 登録日 | 2011-01-30 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural changes that occur upon photolysis of the Fe(II)(a3)-CO complex in the cytochrome ba(3)-oxidase of Thermus thermophilus: A combined X-ray crystallographic and infrared spectral study demonstrates CO binding to Cu(B).
Biochim.Biophys.Acta, 1817, 2012
|
|
7W6L
 
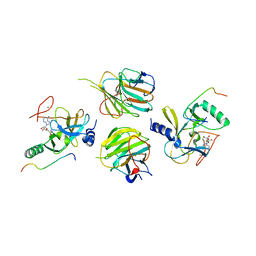 | | The crystal structure of MLL3-RBBP5-ASH2L in complex with H3K4me0 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6A
 
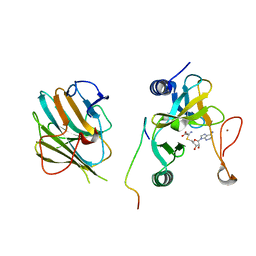 | | Crystal structure of the MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L complex | | 分子名称: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6I
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me1 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W67
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me0 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.194 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6J
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me2 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
8HD0
 
 | | Cell divisome sPG hydrolysis machinery FtsEX-EnvC | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | 著者 | Zhang, Z, Chen, Y. | | 登録日 | 2022-11-03 | | 公開日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Structural insight into the septal peptidoglycan hydrolysis machinery of bacterial cell division
To Be Published
|
|
6K5E
 
 | |
6B0V
 
 | | Crystal Structure of small molecule ARS-107 covalently bound to K-Ras G12C | | 分子名称: | 1-[3-(4-{[(4,5-dichloro-2-hydroxyphenyl)amino]acetyl}piperazin-1-yl)azetidin-1-yl]propan-1-one, CALCIUM ION, GTPase KRas, ... | | 著者 | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | 登録日 | 2017-09-15 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7BWQ
 
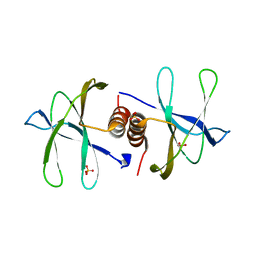 | | Structure of nonstructural protein Nsp9 from SARS-CoV-2 | | 分子名称: | Nsp9, SULFATE ION | | 著者 | Zhang, C, Chen, Y, Li, L, Su, D. | | 登録日 | 2020-04-15 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.954 Å) | | 主引用文献 | Structural basis for the multimerization of nonstructural protein nsp9 from SARS-CoV-2.
Mol Biomed, 1, 2020
|
|
7D42
 
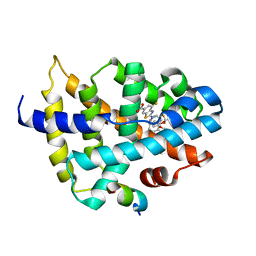 | |
6B0Y
 
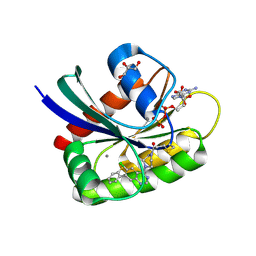 | | Crystal Structure of small molecule ARS-917 covalently bound to K-Ras G12C | | 分子名称: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | 著者 | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | 登録日 | 2017-09-15 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6KUA
 
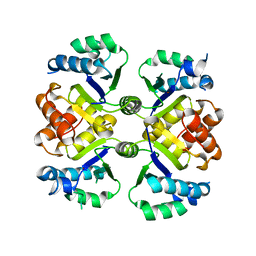 | | Crystal structure of the nicotinamidase SaPncA from Staphylococcus aureus | | 分子名称: | Cysteine hydrolase, ZINC ION | | 著者 | Shang, F, Lan, J, Liu, W, Xu, Y, Chen, Y. | | 登録日 | 2019-08-31 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.104 Å) | | 主引用文献 | Crystal structure of the nicotinamidase SaPncA from Staphylococcus aureus
To Be Published
|
|
3RBT
 
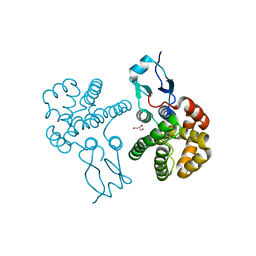 | | Crystal structure of glutathione S-transferase Omega 3 from the silkworm Bombyx mori | | 分子名称: | GLYCEROL, Glutathione transferase o1 | | 著者 | Chen, B.-Y, Ma, X.-X, Tan, X, Yang, J.-P, Zhang, N.-N, Li, W.-F, Chen, Y, Xia, Q, Zhou, C.-Z. | | 登録日 | 2011-03-29 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure-guided activity restoration of the silkworm glutathione transferase Omega GSTO3-3
J.Mol.Biol., 412, 2011
|
|
6L3E
 
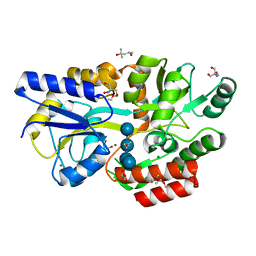 | | Crystal structure of Salmonella enterica sugar-binding protein MalE | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | 著者 | Wang, L, Chen, Y, Liu, W, Lan, J, Shang, F, Xu, Y. | | 登録日 | 2019-10-10 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The crystal structure of Salmonella enterica sugar-binding protein MalE
To Be Published
|
|
7D6C
 
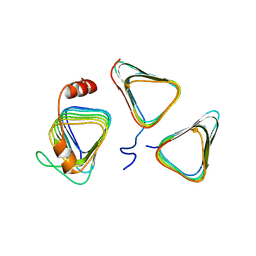 | | Crystal structure of CcmM N-terminal domain in complex with CcmN | | 分子名称: | Carbon dioxide concentrating mechanism protein CcmM, Carboxysome assembly protein CcmN | | 著者 | Sun, H, Cui, N, Han, S.J, Chen, Z.P, Xia, L.Y, Chen, Y, Jiang, Y.L, Zhou, C.Z. | | 登録日 | 2020-09-30 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Complex structure reveals CcmM and CcmN form a heterotrimeric adaptor in beta-carboxysome.
Protein Sci., 30, 2021
|
|
8JUL
 
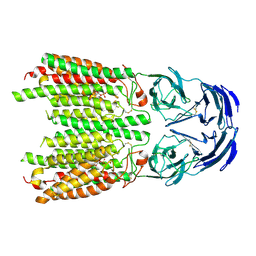 | | Cryo-EM structure of SIDT1 in complex with phosphatidic acid | | 分子名称: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, SID1 transmembrane family member 1, ZINC ION | | 著者 | Sun, C.R, Xu, D, Li, Q, Zhou, C.Z, Chen, Y. | | 登録日 | 2023-06-26 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | Human SIDT1 mediates dsRNA uptake via its phospholipase activity.
Cell Res., 34, 2024
|
|
8JUN
 
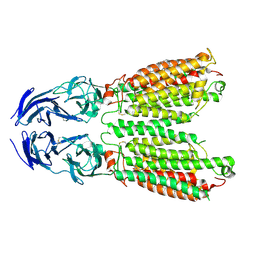 | | Cryo-EM structure of SIDT1 E555Q mutant | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SID1 transmembrane family member 1, ZINC ION | | 著者 | Sun, C.R, Xu, D, Li, Q, Zhou, C.Z, Chen, Y. | | 登録日 | 2023-06-26 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.38 Å) | | 主引用文献 | Human SIDT1 mediates dsRNA uptake via its phospholipase activity.
Cell Res., 34, 2024
|
|
8IQM
 
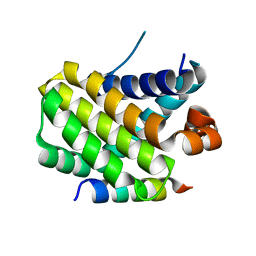 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | 分子名称: | Bcl2 modifying factor, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Wang, H, Guo, M, Wei, H, Chen, Y. | | 登録日 | 2023-03-16 | | 公開日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.967 Å) | | 主引用文献 | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
8IQK
 
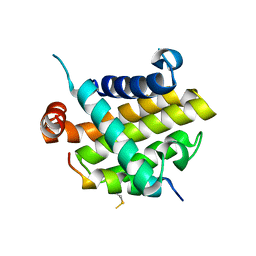 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | 分子名称: | Bcl-2-like protein 1, Bcl-2-modifying factor | | 著者 | Wang, H, Guo, M, Wei, H, Chen, Y. | | 登録日 | 2023-03-16 | | 公開日 | 2023-08-23 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.879 Å) | | 主引用文献 | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
8IQL
 
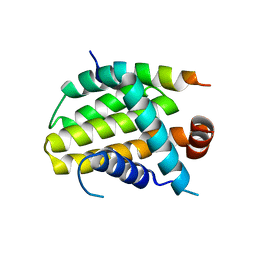 | | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival proteins | | 分子名称: | Apoptosis regulator Bcl-2, Bcl-2-modifying factor | | 著者 | Wang, H, Guo, M, Wei, H, Chen, Y. | | 登録日 | 2023-03-16 | | 公開日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.9577 Å) | | 主引用文献 | Structural basis of the specificity and interaction mechanism of Bmf binding to pro-survival Bcl-2 family proteins.
Comput Struct Biotechnol J, 21, 2023
|
|
7CYE
 
 | | Cryo-EM structure of sodium-dependent bicarbonate transporter SbtA from Synechocystis sp. PCC 6803 | | 分子名称: | Slr1512 protein | | 著者 | Liu, X.Y, Jiang, Y.L, Wang, L, Hou, W.T, Chen, Y, Zhou, C.Z. | | 登録日 | 2020-09-03 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Structures of cyanobacterial bicarbonate transporter SbtA and its complex with PII-like SbtB.
Cell Discov, 7, 2021
|
|
7CYF
 
 | | Cryo-EM structure of bicarbonate transporter SbtA in complex with PII-like signaling protein SbtB from Synechocystis sp. PCC 6803 | | 分子名称: | ADENOSINE MONOPHOSPHATE, Membrane-associated protein slr1513, SODIUM ION, ... | | 著者 | Liu, X.Y, Jiang, Y.L, Wang, L, Hou, W.T, Chen, Y, Zhou, C.Z. | | 登録日 | 2020-09-03 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structures of cyanobacterial bicarbonate transporter SbtA and its complex with PII-like SbtB.
Cell Discov, 7, 2021
|
|
