6PTJ
 
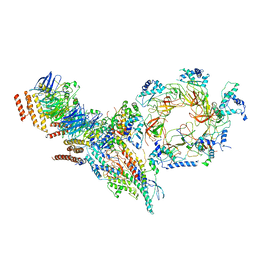 | | Structure of Ctf4 trimer in complex with one CMG helicase | | 分子名称: | Cell division control protein 45, DNA polymerase alpha-binding protein, DNA replication complex GINS protein PSF1, ... | | 著者 | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | 登録日 | 2019-07-15 | | 公開日 | 2019-11-20 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6PSY
 
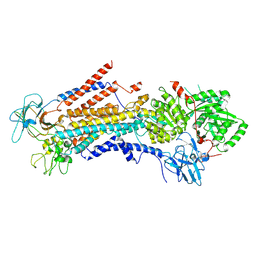 | | Cryo-EM structure of S. cerevisiae Drs2p-Cdc50p in the autoinhibited apo form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell division control protein 50, ... | | 著者 | Bai, L, Li, H. | | 登録日 | 2019-07-14 | | 公開日 | 2019-09-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p-Cdc50p.
Nat Commun, 10, 2019
|
|
5H3O
 
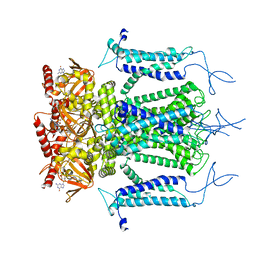 | | Structure of a eukaryotic cyclic nucleotide-gated channel | | 分子名称: | CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel, SODIUM ION | | 著者 | Li, M, Zhou, X, Wang, S, Michailidis, I, Gong, Y, Su, D, Li, H, Li, X, Yang, J. | | 登録日 | 2016-10-26 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of a eukaryotic cyclic-nucleotide-gated channel.
Nature, 542, 2017
|
|
6PTN
 
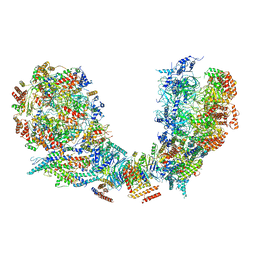 | | Structure of Ctf4 trimer in complex with two CMG helicases | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | 著者 | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | 登録日 | 2019-07-16 | | 公開日 | 2019-11-20 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6PSX
 
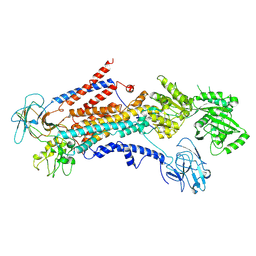 | | Cryo-EM structure of S. cerevisiae Drs2p-Cdc50p in the PI4P-activated form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell division control protein 50, ... | | 著者 | Bai, L, Li, H. | | 登録日 | 2019-07-14 | | 公開日 | 2019-09-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p-Cdc50p.
Nat Commun, 10, 2019
|
|
6NSE
 
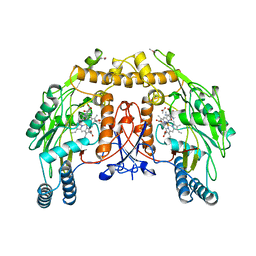 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, H4B-FREE, CANAVANINE COMPLEX | | 分子名称: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | 著者 | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | 登録日 | 1999-01-13 | | 公開日 | 2002-05-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal Structures of the Heme Domain of Bovine Endothelial Nitric Oxide Synthase Complexed with Arginine Analogues
To be Published
|
|
6U0M
 
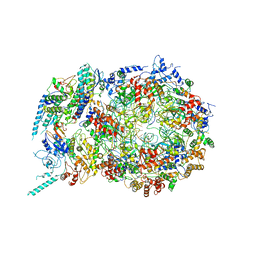 | | Structure of the S. cerevisiae replicative helicase CMG in complex with a forked DNA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA (15-MER), ... | | 著者 | Yuan, Z, Georgescu, R, Bai, L, Zhang, D, O'Donnell, M, Li, H. | | 登録日 | 2019-08-14 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | DNA unwinding mechanism of a eukaryotic replicative CMG helicase.
Nat Commun, 11, 2020
|
|
7T30
 
 | |
7T2R
 
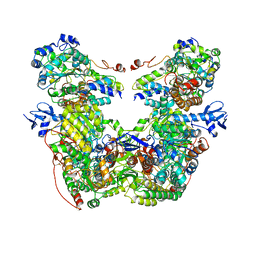 | |
2HX4
 
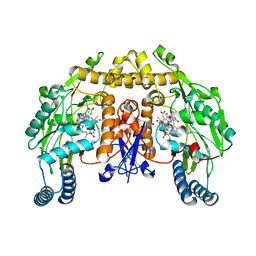 | | Rat nNOS heme domain complexed with 4-N-(Nw-nitro-L-argininyl)-trans-4-hydroxyamino-L-proline amide | | 分子名称: | (4R)-4-(HYDROXY{N~5~-[IMINO(NITROAMINO)METHYL]-L-ORNITHYL}AMINO)-L-PROLINAMIDE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | 著者 | Igarashi, J, Li, H, Poulos, T.L. | | 登録日 | 2006-08-02 | | 公開日 | 2007-04-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure-Based Design and Synthesis of N(omega)-Nitro-l-Arginine-Containing Peptidomimetics as Selective Inhibitors of Neuronal Nitric Oxide Synthase. Displacement of the Heme Structural Water.
J.Med.Chem., 50, 2007
|
|
2HX2
 
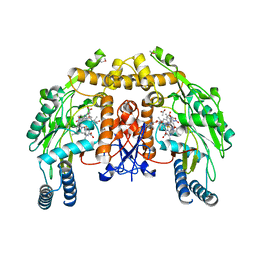 | | Bovine eNOS heme domain complexed with (4S)-N-{4-Amino-5-[(2-aminoethyl)-hydroxyamino]-pentyl}-N'-nitroguanidine | | 分子名称: | (4S)-N-{4-AMINO-5-[(2-AMINOETHYL)(HYDROXYAMINO]-PENTYL}-N'-NITROGUANIDINE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | 著者 | Igarashi, J, Li, H, Poulos, T.L. | | 登録日 | 2006-08-02 | | 公開日 | 2007-04-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-Based Design and Synthesis of N(omega)-Nitro-l-Arginine-Containing Peptidomimetics as Selective Inhibitors of Neuronal Nitric Oxide Synthase. Displacement of the Heme Structural Water.
J.Med.Chem., 50, 2007
|
|
2HX3
 
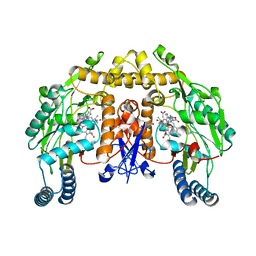 | | Rat nNOS heme domain complexed with (4S)-N-{4-Amino-5-[(2-aminoethyl)-hydroxyamino]-pentyl}-N'-nitroguanidine | | 分子名称: | (4S)-N-{4-AMINO-5-[(2-AMINOETHYL)(HYDROXYAMINO]-PENTYL}-N'-NITROGUANIDINE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | 著者 | Igarashi, J, Li, H, Poulos, T.L. | | 登録日 | 2006-08-02 | | 公開日 | 2007-04-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Based Design and Synthesis of N(omega)-Nitro-l-Arginine-Containing Peptidomimetics as Selective Inhibitors of Neuronal Nitric Oxide Synthase. Displacement of the Heme Structural Water.
J.Med.Chem., 50, 2007
|
|
1S6V
 
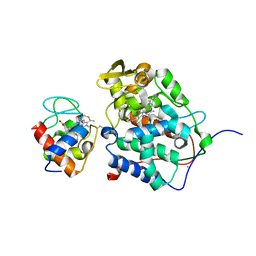 | | Structure of a cytochrome c peroxidase-cytochrome c site specific cross-link | | 分子名称: | Cytochrome c peroxidase, mitochondrial, Cytochrome c, ... | | 著者 | Guo, M, Bhaskar, B, Li, H, Barrows, T.P, Poulos, T.L. | | 登録日 | 2004-01-27 | | 公開日 | 2004-04-27 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystal structure and characterization of a cytochrome c peroxidase-cytochrome c site-specific cross-link
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3JWX
 
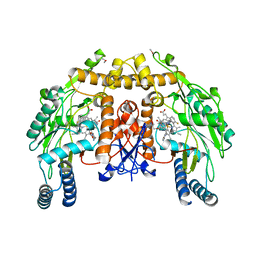 | | Structure of endothelial nitric oxide synthase heme domain complexed with N1-[(3'R,4'R)-4'-((6"-amino-4"-methylpyridin-2"-yl)methyl)pyrrolidin-3'-yl]-N2-(3'-fluorophenethyl)ethane-1,2-diamine tetrahydrochloride | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLIC ACID, ... | | 著者 | Delker, S.L, Li, H, Poulos, T.L. | | 登録日 | 2009-09-18 | | 公開日 | 2010-05-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
3JX6
 
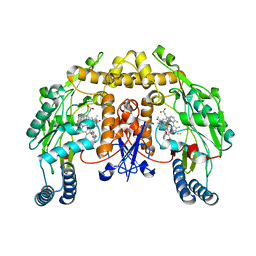 | | Structure of neuronal nitric oxide synthase D597N/M336V/Y706A mutant heme domain complexed with N1-[(3' R,4' R)-4'-((6"-amino-4"-methylpyridin-2"-yl)methyl)pyrrolidin-3'-yl]-N2-(3'-fluorophenethyl)ethane-1,2-diamine tetrahydrochloride | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4R)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | 著者 | Delker, S.L, Li, H, Poulos, T.L. | | 登録日 | 2009-09-18 | | 公開日 | 2010-05-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
3P2M
 
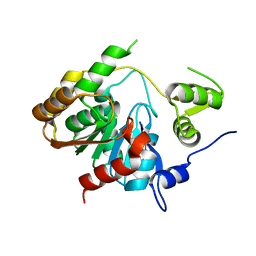 | | Crystal Structure of a Novel Esterase Rv0045c from Mycobacterium tuberculosis | | 分子名称: | POSSIBLE HYDROLASE | | 著者 | Zheng, X.D, Guo, J, Xu, L, Li, H, Zhang, D, Zhang, K, Sun, F, Wen, T, Liu, S, Pang, H. | | 登録日 | 2010-10-03 | | 公開日 | 2011-07-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of a Novel Esterase Rv0045c from Mycobacterium tuberculosis
Plos One, 6, 2011
|
|
8G6F
 
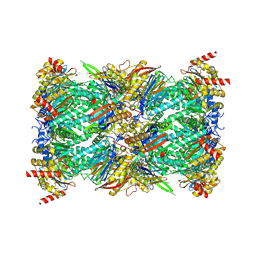 | |
8G6E
 
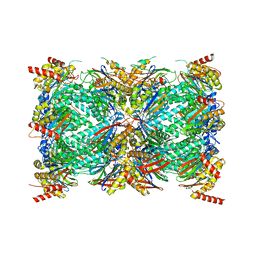 | | Structure of the Plasmodium falciparum 20S proteasome complexed with inhibitor TDI-8304 | | 分子名称: | (7S,10S,13S)-N-cyclopentyl-10-[2-(morpholin-4-yl)ethyl]-9,12-dioxo-13-(2-oxopyrrolidin-1-yl)-2-oxa-8,11-diazabicyclo[13.3.1]nonadeca-1(19),15,17-triene-7-carboxamide, Proteasome subunit alpha type, Proteasome subunit alpha type-1, ... | | 著者 | Hsu, H.-C, Li, H. | | 登録日 | 2023-02-15 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (2.18 Å) | | 主引用文献 | Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors.
Nat Commun, 14, 2023
|
|
3JWV
 
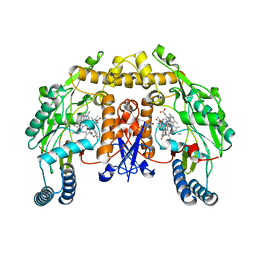 | | Structure of rat neuronal nitric oxide synthase R349A mutant heme domain in complex with N1-{(3'S,4'R)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4R)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | 著者 | Delker, S.L, Li, H, Poulos, T.L. | | 登録日 | 2009-09-18 | | 公開日 | 2010-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
3NLD
 
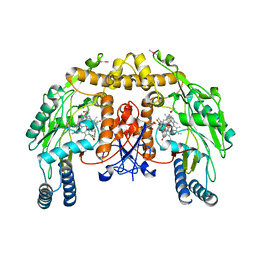 | | Structure of endothelial nitric oxide synthase heme domain complexed with 6-{{(3'S,4'S)-3'-[2"-(3'''-fluorophenethylamino)ethoxy]pyrrolidin-4'-yl}methyl}-4-methylpyridin-2-amine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3S,4S)-4-(2-{[2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | 著者 | Ji, H, Delker, S.L, Li, H, Martasek, P, Roman, L, Poulos, T.L, Silverman, R.B. | | 登録日 | 2010-06-21 | | 公開日 | 2010-11-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.285 Å) | | 主引用文献 | Exploration of the Active Site of Neuronal Nitric Oxide Synthase by the Design and Synthesis of Pyrrolidinomethyl 2-Aminopyridine Derivatives.
J.Med.Chem., 53, 2010
|
|
3DQS
 
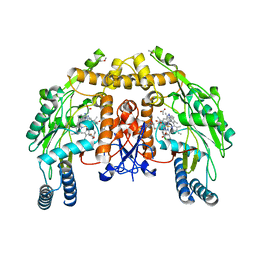 | | Structure of endothelial NOS heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(4'-chlorobenzyl)ethane-1,2-diamine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | 著者 | Igarashi, J, Li, H, Poulos, T.L. | | 登録日 | 2008-07-09 | | 公開日 | 2009-03-31 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
3JWS
 
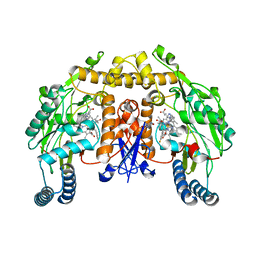 | | Structure of neuronal nitric oxide synthase R349A mutant heme domain complexed with N1-[(3' S,4'S)-4'-((6"-amino-4"-methylpyridin-2"-yl)methyl)pyrrolidin-3'-yl]-N2-(3'-fluorophenethyl)ethane-1,2-diamine tetrahydrochloride | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | 著者 | Delker, S.L, Li, H, Poulos, T.L. | | 登録日 | 2009-09-18 | | 公開日 | 2010-05-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
3JX2
 
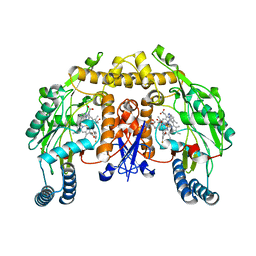 | | Structure of rat neuronal nitric oxide synthase D597N/M336V mutant heme domain in complex with N1-{(3'S,4'S)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, GLYCEROL, ... | | 著者 | Delker, S.L, Li, H, Poulos, T.L. | | 登録日 | 2009-09-18 | | 公開日 | 2010-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
3NLF
 
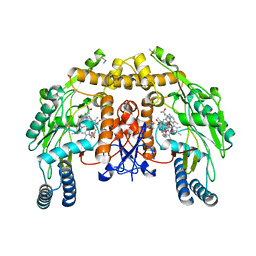 | | Structure of endothelial nitric oxide synthase heme domain complexed with 6-{{(3'R,4'S)-3'-[2"-(3'''-fluorophenethylamino)ethoxy]pyrrolidin-4'-yl}methyl}-4-methylpyridin-2-amine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3S,4R)-4-(2-{[2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | 著者 | Ji, H, Delker, S.L, Li, H, Martasek, P, Roman, L, Poulos, T.L, Silverman, R.B. | | 登録日 | 2010-06-21 | | 公開日 | 2010-11-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Exploration of the Active Site of Neuronal Nitric Oxide Synthase by the Design and Synthesis of Pyrrolidinomethyl 2-Aminopyridine Derivatives.
J.Med.Chem., 53, 2010
|
|
3JWZ
 
 | | Structure of endothelial nitric oxide synthase heme domain complexed with N1-[(3' S,4' R)-4'-((6"-amino-4"-methylpyridin-2"-yl)methyl)pyrrolidin-3'-yl]-N2-(3'-fluorophenethyl)ethane-1,2-diamine tetrahydrochloride | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLIC ACID, ... | | 著者 | Delker, S.L, Li, H, Poulos, T.L. | | 登録日 | 2009-09-18 | | 公開日 | 2010-05-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
