4LWI
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ6 | | 分子名称: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl}cyclopropanecarboxamide | | 著者 | Li, J, Shi, F, Xiong, B, He, J. | | 登録日 | 2013-07-27 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
8XEP
 
 | | Crystal structure of a Legionella pneumophila type IV effector in complex with ubiquitin | | 分子名称: | SULFATE ION, Type IV effector MavL, Ubiquitin | | 著者 | Tan, J.X, Wang, X.F, Zhou, Y, Zhu, Y.Q. | | 登録日 | 2023-12-12 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Legionella effector LnaB is a phosphoryl-AMPylase that impairs phosphosignalling.
Nature, 2024
|
|
5CNI
 
 | | mGlu2 with Glutamate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | 著者 | Clawson, D.K, Atwell, S, Monn, J.A. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-02-15 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Synthesis and Pharmacological Characterization of C4-(Thiotriazolyl)-substituted-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1R,2S,4R,5R,6R)-2-Amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2812223), a Highly Potent, Functionally Selective mGlu2 Receptor Agonist.
J.Med.Chem., 58, 2015
|
|
5CNM
 
 | | mGluR3 complexed with glutamate analog | | 分子名称: | (1R,2S,4R,5R,6R)-2-amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Monn, J.A, Clawson, D.K, McKinzie, D. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-09 | | 最終更新日 | 2015-10-07 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Synthesis and Pharmacological Characterization of C4-(Thiotriazolyl)-substituted-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1R,2S,4R,5R,6R)-2-Amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2812223), a Highly Potent, Functionally Selective mGlu2 Receptor Agonist.
J.Med.Chem., 58, 2015
|
|
5CH8
 
 | | Crystal structure of MDLA N225Q mutant form Penicillium cyclopium | | 分子名称: | CHLORIDE ION, GLYCEROL, Mono- and diacylglycerol lipase, ... | | 著者 | Xu, J, Xu, H, Hou, S, Liu, J. | | 登録日 | 2015-07-10 | | 公開日 | 2016-04-20 | | 最終更新日 | 2017-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Lipase-Driven Epoxidation Is A Two-Stage Synergistic Process
ChemistrySelect, 4, 2016
|
|
3NI2
 
 | | Crystal structures and enzymatic mechanisms of a Populus tomentosa 4-coumarate:CoA ligase | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-coumarate:CoA ligase, 5'-O-{(S)-hydroxy[3-(4-hydroxyphenyl)propoxy]phosphoryl}adenosine | | 著者 | Hu, Y, Yin, L, Gai, Y, Wang, X.X, Wang, D.C. | | 登録日 | 2010-06-14 | | 公開日 | 2010-09-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of a Populus tomentosa 4-coumarate:CoA ligase shed light on its enzymatic mechanisms
Plant Cell, 22, 2010
|
|
3EIJ
 
 | | Crystal structure of Pdcd4 | | 分子名称: | Programmed cell death protein 4 | | 著者 | Loh, P.G. | | 登録日 | 2008-09-16 | | 公開日 | 2009-02-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for translational inhibition by the tumour suppressor Pdcd4
Embo J., 28, 2009
|
|
3EIQ
 
 | | Crystal structure of Pdcd4-eIF4A | | 分子名称: | Eukaryotic initiation factor 4A-I, Programmed cell death protein 4 | | 著者 | Loh, P.G, Cheng, Z, Song, H. | | 登録日 | 2008-09-17 | | 公開日 | 2009-02-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural basis for translational inhibition by the tumour suppressor Pdcd4
Embo J., 28, 2009
|
|
6VET
 
 | | Human insulin analog: [GluB10,HisA8,ArgA9,TyrB20]-DOI | | 分子名称: | Insulin A chain, Insulin B chain | | 著者 | Menting, J.G, Chou, D.H.-C, Lawrence, M.C, Xiong, X. | | 登録日 | 2020-01-02 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | A structurally minimized yet fully active insulin based on cone-snail venom insulin principles.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VEQ
 
 | | Con-Ins G1 in complex with the human insulin microreceptor in turn in complex with Fv 83-7 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Con-Ins G1 B chain, Con-Ins G1a A chain, ... | | 著者 | Menting, J.G, Chou, D.H.-C, Lawrence, M.C, Xiong, X. | | 登録日 | 2020-01-02 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | A structurally minimized yet fully active insulin based on cone-snail venom insulin principles.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8X4Z
 
 | | BA.2.86 Spike Trimer with ins483V mutation (3 RBD down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8X56
 
 | | BA.2.86 Spike Trimer with T356K mutation (1 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8WHZ
 
 | | BA.2.86 RBD in complex with hACE2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-09-23 | | 公開日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8X55
 
 | | BA.2.86 Spike Trimer with T356K mutation (3 RBD down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8WHU
 
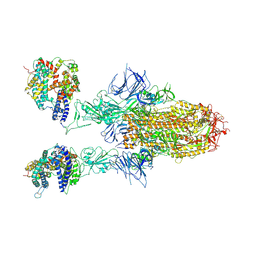 | | Spike Trimer of BA.2.86 in complex with two hACE2s | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-09-23 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8X5R
 
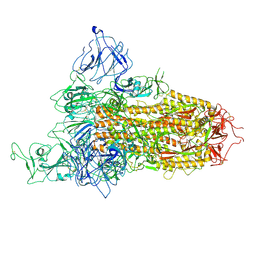 | | SARS-CoV-2 BA.2.75 Spike with K356T mutation (1 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-17 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8X50
 
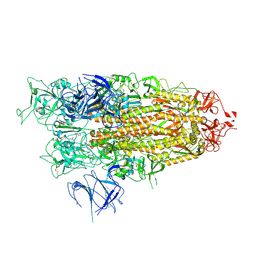 | | BA.2.86 Spike Trimer with ins483V mutation (1 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-16 | | 公開日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.82 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8X4H
 
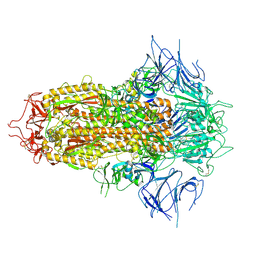 | | SARS-CoV-2 JN.1 Spike | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-15 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8X5Q
 
 | | SARS-CoV-2 BA.2.75 Spike with K356T mutation (3 RBD down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-11-17 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8WHW
 
 | | Spike Trimer of BA.2.86 with single RBD up | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-09-23 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8WHV
 
 | | Spike Trimer of BA.2.86 with three RBDs down | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-09-23 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8WHS
 
 | | Spike Trimer of BA.2.86 in complex with one hACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | 著者 | Yue, C, Liu, P. | | 登録日 | 2023-09-23 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
8XUS
 
 | | JN.1 Spike Trimer in complex with heparan sulfate | | 分子名称: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yue, C, Liu, P. | | 登録日 | 2024-01-14 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity
Natl Sci Rev, 2024
|
|
4O9S
 
 | | Crystal structure of Retinol-Binding Protein 4 (RBP4)in complex with a non-retinoid ligand | | 分子名称: | 1,2-ETHANEDIOL, 1-[4-(7-thia-9,11-diazatricyclo[6.4.0.0^{2,6}]dodeca-1(12),2(6),8,10-tetraen-12-yl)piperazin-1-yl]-2-[2-(trifluoromethyl)phenyl]ethanone, CHLORIDE ION, ... | | 著者 | Wang, Z, Johnstone, S, Walker, N.P. | | 登録日 | 2014-01-02 | | 公開日 | 2014-07-02 | | 最終更新日 | 2020-02-19 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-assisted discovery of the first non-retinoid ligands for Retinol-Binding Protein 4.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4PSQ
 
 | | Crystal Structure of Retinol-Binding Protein 4 (RBP4) in complex with a non-retinoid ligand | | 分子名称: | (1-benzyl-1H-imidazol-4-yl)[4-(2-chlorophenyl)piperazin-1-yl]methanone, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | 著者 | Wang, Z, Johnstone, S, Walker, N. | | 登録日 | 2014-03-07 | | 公開日 | 2014-07-02 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-assisted discovery of the first non-retinoid ligands for Retinol-Binding Protein 4.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
